User Guide
Contents
User Guide#
Table of contents:
Reading genetic data#
Installation#
Sgkit can read standard genetic file formats, including VCF, PLINK, and BGEN.
If sgkit has been installed using conda, support for reading BGEN and PLINK is included, but VCF is not because there is no Windows support for cyvcf2, the library we use for reading VCF data. If you are using Linux or a Mac, please install cyvcf2 using the following to enable VCF support:
$ conda install -c bioconda cyvcf2
If sgkit has been installed using pip, then support for reading these formats is not included, and requires additional dependencies, which can be installed as an “extra” feature using pip, as follows.
To install sgkit with BGEN support:
$ pip install 'sgkit[bgen]'
To install sgkit with PLINK support:
$ pip install 'sgkit[plink]'
To install sgkit with VCF support:
$ pip install 'sgkit[vcf]'
Converting genetic data to Zarr#
There are broadly two ways of loading genetic file format X for use in sgkit:
a
read_Xfunction to read the file directlya
X_to_zarrfunction to convert to Zarr first
Which one to use depends on the size of the input, as well as the file format (not all file formats have both options).
Generally speaking, the read_X functions are convenience functions that are suitable
for small input files, or where the cost of conversion is small. The X_to_zarr functions
are more appropriate for large datasets, since the expensive conversion step need only be
paid once.
After converting a file to Zarr format, it can be loaded with sgkit’s sgkit.load_dataset().
This is like Xarray’s xarray.open_zarr() function, with some useful defaults applied for you.
Similarly, the function sgkit.save_dataset() can be used to save the dataset in Zarr format.
This can be used to save a newly-loaded dataset, or a dataset with new variables that are the
result of running genetic methods on the data, so it can be loaded again later.
By default, Zarr datasets created by sgkit (and Xarray) have consolidated metadata, which makes opening the dataset faster.
BGEN#
The sgkit.io.bgen.read_bgen() function loads a single BGEN dataset as Dask
arrays within an xarray.Dataset from a bgen file.
The sgkit.io.bgen.bgen_to_zarr() function converts a bgen file to Zarr
files stored in sgkit’s Xarray data representation, which can then be opened
as a xarray.Dataset.
PLINK#
The sgkit.io.plink.read_plink() function loads a single PLINK dataset as Dask
arrays within an xarray.Dataset from bed, bim, and fam files.
VCF#
The sgkit.io.vcf.vcf_to_zarr() function converts one or more VCF files to
Zarr files stored in sgkit’s Xarray data representation, which can then be opened
as a xarray.Dataset.
See Reading VCF for installation instructions, and details on using VCF in sgkit.
Working with cloud-native data#
TODO: Show how to read/write Zarr (and VCF?) data in cloud storage
Datasets#
Genetic variables#
Most Genetic methods in sgkit operate on a few variables in an Xarray dataset. Variables have default names, so you can usually just pass in the dataset, but it’s also possible to use different variable names.
In [1]: import sgkit as sg
In [2]: ds = sg.simulate_genotype_call_dataset(n_variant=100, n_sample=50, missing_pct=.1)
In [3]: ds = ds[['variant_allele', 'call_genotype']]
In [4]: ds
Out[4]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2, samples: 50, ploidy: 2)
Dimensions without coordinates: variants, alleles, samples, ploidy
Data variables:
variant_allele (variants, alleles) |S1 b'T' b'C' b'C' ... b'T' b'T' b'A'
call_genotype (variants, samples, ploidy) int8 0 0 1 0 1 0 ... 0 1 0 1 0 0
Attributes:
contigs: [0]
source: sgkit-0.4.1.dev20+g839eb9a9
# Use the default variable (call_genotype)
In [5]: sg.count_call_alleles(ds).call_allele_count
Out[5]:
<xarray.DataArray 'call_allele_count' (variants: 100, samples: 50, alleles: 2)>
dask.array<count_alleles, shape=(100, 50, 2), dtype=uint8, chunksize=(100, 50, 2), chunktype=numpy.ndarray>
Dimensions without coordinates: variants, samples, alleles
Attributes:
comment: Allele counts. With shape (variants, samples, alleles) and valu...
# Create a copy of the call_genotype variable, and use that to compute counts
# (More realistically, this variable would be created from another computation or input.)
In [6]: ds["my_call_genotype"] = ds["call_genotype"]
In [7]: sg.count_call_alleles(ds, call_genotype="my_call_genotype").call_allele_count
Out[7]:
<xarray.DataArray 'call_allele_count' (variants: 100, samples: 50, alleles: 2)>
dask.array<count_alleles, shape=(100, 50, 2), dtype=uint8, chunksize=(100, 50, 2), chunktype=numpy.ndarray>
Dimensions without coordinates: variants, samples, alleles
Attributes:
comment: Allele counts. With shape (variants, samples, alleles) and valu...
For a full list of variables and their default names, see Variables.
Methods declare the variables that they use directly. If the variable exists in the dataset, then it will be used for the computation.
If the variable doesn’t exist in the dataset, then it will be computed if the variable name is
the default one. For example, sgkit.count_variant_alleles() declares
call_allele_count as a variable it needs to perform its computation.
If the dataset doesn’t contain call_allele_count, then the method will
call sgkit.count_call_alleles() to populate it, before running its own computation.
# The following will create call_allele_count and variant_allele_count
In [8]: sg.count_variant_alleles(ds)
Out[8]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2, samples: 50, ploidy: 2)
Dimensions without coordinates: variants, alleles, samples, ploidy
Data variables:
variant_allele_count (variants, alleles) uint64 dask.array<chunksize=(100, 2), meta=np.ndarray>
call_allele_count (variants, samples, alleles) uint8 dask.array<chunksize=(100, 50, 2), meta=np.ndarray>
variant_allele (variants, alleles) |S1 b'T' b'C' b'C' ... b'T' b'A'
call_genotype (variants, samples, ploidy) int8 0 0 1 0 1 ... 0 1 0 0
my_call_genotype (variants, samples, ploidy) int8 0 0 1 0 1 ... 0 1 0 0
Attributes:
contigs: [0]
source: sgkit-0.4.1.dev20+g839eb9a9
If however a non-default variable name is used and it doesn’t exist in the dataset, then the intermediate variable is not populated, and an error is raised, since sgkit expects the user to have created the variable in that case.
In [9]: sg.count_variant_alleles(ds, call_allele_count="my_call_allele_count")
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-9-a02760c2a541> in <cell line: 1>()
----> 1 sg.count_variant_alleles(ds, call_allele_count="my_call_allele_count")
~/workspace/sgkit/sgkit/stats/aggregation.py in count_variant_alleles(ds, call_allele_count, merge)
218 [4, 0]], dtype=uint64)
219 """
--> 220 ds = define_variable_if_absent(
221 ds, variables.call_allele_count, call_allele_count, count_call_alleles
222 )
~/workspace/sgkit/sgkit/utils.py in define_variable_if_absent(ds, default_variable_name, variable_name, func)
190 return ds
191 if variable_name != default_variable_name:
--> 192 raise ValueError(
193 f"Variable '{variable_name}' with non-default name is missing and will not be automatically defined."
194 )
ValueError: Variable 'my_call_allele_count' with non-default name is missing and will not be automatically defined.
There are also some variables that cannot be automatically defined, such as call_genotype,
since it can’t be computed from other data.
Dataset merge behavior#
Generally, method functions in sgkit compute some new variables based on the input dataset, then return a new output dataset that consists of the input dataset plus the new computed variables. The input dataset is unchanged.
This behavior can be controlled using the merge parameter. If set to True
(the default), then the function will merge the input dataset and the computed
output variables into a single dataset. Output variables will overwrite any
input variables with the same name, and a warning will be issued in this case.
If False, the function will return only the computed output variables.
Examples:
In [10]: import sgkit as sg
In [11]: ds = sg.simulate_genotype_call_dataset(n_variant=100, n_sample=50, missing_pct=.1)
In [12]: ds = ds[['variant_allele', 'call_genotype']]
In [13]: ds
Out[13]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2, samples: 50, ploidy: 2)
Dimensions without coordinates: variants, alleles, samples, ploidy
Data variables:
variant_allele (variants, alleles) |S1 b'T' b'C' b'C' ... b'T' b'T' b'A'
call_genotype (variants, samples, ploidy) int8 0 0 1 0 1 0 ... 0 1 0 1 0 0
Attributes:
contigs: [0]
source: sgkit-0.4.1.dev20+g839eb9a9
# By default, new variables are merged into a copy of the provided dataset
In [14]: ds = sg.count_variant_alleles(ds)
In [15]: ds
Out[15]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2, samples: 50, ploidy: 2)
Dimensions without coordinates: variants, alleles, samples, ploidy
Data variables:
variant_allele_count (variants, alleles) uint64 dask.array<chunksize=(100, 2), meta=np.ndarray>
call_allele_count (variants, samples, alleles) uint8 dask.array<chunksize=(100, 50, 2), meta=np.ndarray>
variant_allele (variants, alleles) |S1 b'T' b'C' b'C' ... b'T' b'A'
call_genotype (variants, samples, ploidy) int8 0 0 1 0 1 ... 0 1 0 0
Attributes:
contigs: [0]
source: sgkit-0.4.1.dev20+g839eb9a9
# If an existing variable would be re-defined, a warning is thrown
In [16]: import warnings
In [17]: ds = sg.count_variant_alleles(ds)
In [18]: with warnings.catch_warnings(record=True) as w:
....: ds = sg.count_variant_alleles(ds)
....: print(f"{w[0].category.__name__}: {w[0].message}")
....:
MergeWarning: The following variables in the input dataset will be replaced in the output: variant_allele_count
# New variables can also be returned in their own dataset
In [19]: sg.count_variant_alleles(ds, merge=False)
Out[19]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2)
Dimensions without coordinates: variants, alleles
Data variables:
variant_allele_count (variants, alleles) uint64 dask.array<chunksize=(100, 2), meta=np.ndarray>
# This can be useful for merging multiple datasets manually
In [20]: ds.merge(sg.count_variant_alleles(ds, merge=False))
Out[20]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2, samples: 50, ploidy: 2)
Dimensions without coordinates: variants, alleles, samples, ploidy
Data variables:
variant_allele_count (variants, alleles) uint64 dask.array<chunksize=(100, 2), meta=np.ndarray>
call_allele_count (variants, samples, alleles) uint8 dask.array<chunksize=(100, 50, 2), meta=np.ndarray>
variant_allele (variants, alleles) |S1 b'T' b'C' b'C' ... b'T' b'A'
call_genotype (variants, samples, ploidy) int8 0 0 1 0 1 ... 0 1 0 0
Attributes:
contigs: [0]
source: sgkit-0.4.1.dev20+g839eb9a9
Merge can be used to rename output variables too.
In [21]: import sgkit as sg
In [22]: ds = sg.simulate_genotype_call_dataset(n_variant=100, n_sample=50, missing_pct=.1)
In [23]: ds = ds[['variant_allele', 'call_genotype']]
In [24]: ds.merge(sg.count_variant_alleles(ds, merge=False).rename(variant_allele_count="my_variant_allele_count"))
Out[24]:
<xarray.Dataset>
Dimensions: (variants: 100, alleles: 2, samples: 50, ploidy: 2)
Dimensions without coordinates: variants, alleles, samples, ploidy
Data variables:
variant_allele (variants, alleles) |S1 b'T' b'C' ... b'T' b'A'
call_genotype (variants, samples, ploidy) int8 0 0 1 0 ... 1 0 0
my_variant_allele_count (variants, alleles) uint64 dask.array<chunksize=(100, 2), meta=np.ndarray>
Attributes:
contigs: [0]
source: sgkit-0.4.1.dev20+g839eb9a9
Note that there is a limitation where intermediate variables (call_allele_count in this case)
are not returned if merge=False. See https://github.com/pystatgen/sgkit/issues/405.
Custom naming conventions#
TODO: Show to use a custom naming convention via Xarray renaming features.
Adding custom data to a Dataset#
TODO: Show how something like sample metadata can be joined to an existing Xarray dataset. Also briefly explain indexing and uniqueness within Xarray/Pandas, since this is critical for understanding joins.
Methods#
Custom Computations#
TODO: Finish explaining how Numba works and how users might apply it
Here is an example that demonstrates an alt allele count:
In [25]: import numba
In [26]: import sgkit as sg
In [27]: import numpy as np
In [28]: ds = sg.simulate_genotype_call_dataset(5, 3, missing_pct=.2)
In [29]: def alt_allele_count(gt):
....: out = np.full(gt.shape[:2], -1, dtype=np.int64)
....: for i, j in np.ndindex(*out.shape):
....: if np.all(gt[i, j] >= 0):
....: out[i, j] = np.sum(gt[i, j] > 0)
....: return out
....:
In [30]: numba.njit(alt_allele_count)(ds.call_genotype.values)
Out[30]:
array([[ 0, 1, 1],
[ 1, 1, -1],
[-1, 1, 2],
[ 0, -1, -1],
[ 1, 2, 2]])
PCA#
TODO: Describe the upstream tools for PCA (i.e. those in dask-ml/scikit-learn)
Deployment#
Deploying sgkit on a cluster#
TODO: Create a tutorial on running sgkit at scale
Using GPUs#
TODO: Show CuPy examples
Troubleshooting#
Monitoring operations#
The simplest way to monitor operations when running sgkit on a single host is to use Dask local diagnostics.
As an example, this code shows how to track the progress of a single sgkit function:
In [31]: import sgkit as sg
In [32]: from dask.diagnostics import ProgressBar
In [33]: ds = sg.simulate_genotype_call_dataset(n_variant=100, n_sample=50, missing_pct=.1)
In [34]: with ProgressBar():
....: ac = sg.count_variant_alleles(ds).variant_allele_count.compute()
....:
[ ] | 0% Completed | 0.0s
[########################################] | 100% Completed | 0.1s
In [35]: ac[:5]
Out[35]:
<xarray.DataArray 'variant_allele_count' (variants: 5, alleles: 2)>
array([[50, 40],
[40, 52],
[48, 45],
[37, 51],
[45, 46]], dtype=uint64)
Dimensions without coordinates: variants, alleles
Attributes:
comment: Variant allele counts. With shape (variants, alleles) and value...
Monitoring resource utilization with ResourceProfiler and profiling task streams with Profiler are other commonly used local diagnostics.
For similar monitoring in a distributed cluster, see Dask distributed diagnostics.
Visualizing computations#
Dask allows you to visualize the task graph of a computation before running it, which can be handy when trying to understand where the bottlenecks are.
In most cases the number of tasks is too large to visualize, so it’s useful to restrict the graph just a few chunks, as shown in this example.
In [36]: import sgkit as sg
In [37]: ds = sg.simulate_genotype_call_dataset(n_variant=100, n_sample=50, missing_pct=.1)
# Rechunk to illustrate multiple tasks
In [38]: ds = ds.chunk({"variants": 25, "samples": 25})
In [39]: counts = sg.count_call_alleles(ds).call_allele_count.data
# Restrict to first 3 chunks in variants dimension
In [40]: counts = counts[:3*counts.chunksize[0],...]
In [41]: counts.visualize(optimize_graph=True)
Out[41]: <IPython.core.display.Image object>
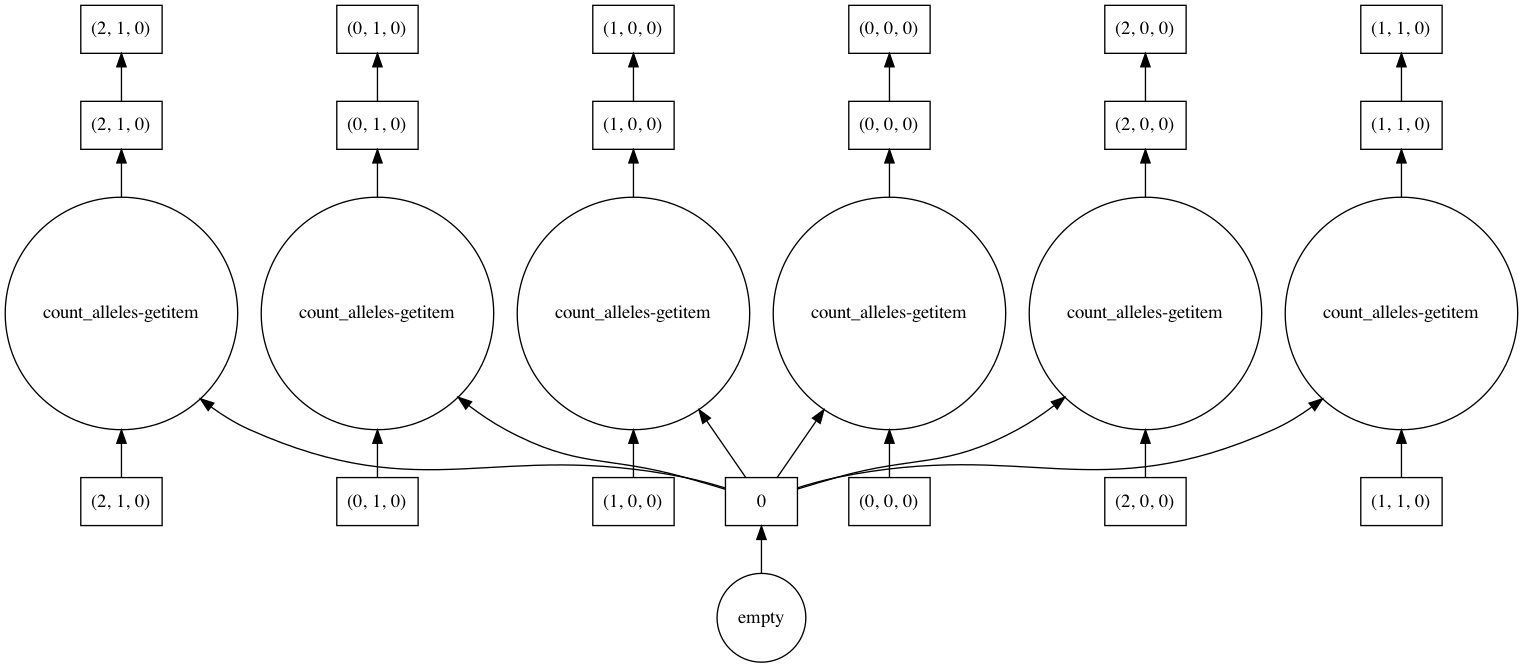
By passing keyword arguments to visualize we can see the order tasks will run in:
# Graph where colors indicate task ordering
In [42]: counts.visualize(filename="order", optimize_graph=True, color="order", cmap="autumn", node_attr={"penwidth": "4"})
Out[42]: <IPython.core.display.Image object>
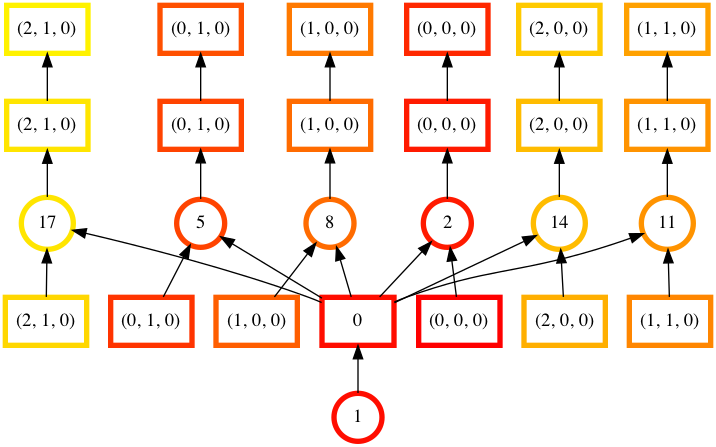
Task order number is shown in circular boxes, colored from red to yellow.
