GWAS Tutorial#
This notebook is an sgkit port of Hail’s GWAS Tutorial, which demonstrates how to run a genome-wide SNP association test. Readers are encouraged to read the Hail tutorial alongside this one for more background, and to see the motivation behind some of the steps.
Note that some of the results do not exactly match the output from Hail. Also, since sgkit is still a 0.x release, its API is still subject to non-backwards compatible changes.
import sgkit as sg
Before using sgkit, we import some standard Python libraries and set the Xarray display options to not show all the attributes in a dataset by default.
import numpy as np
import pandas as pd
import xarray as xr
xr.set_options(display_expand_attrs=False, display_expand_data_vars=True);
Download public 1000 Genomes data#
We use the same small (20MB) portion of the public 1000 Genomes data that Hail uses.
First, download the file locally:
from pathlib import Path
import requests
if not Path("1kg.vcf.bgz").exists():
response = requests.get("https://storage.googleapis.com/sgkit-data/tutorial/1kg.vcf.bgz")
with open("1kg.vcf.bgz", "wb") as f:
f.write(response.content)
if not Path("1kg.vcf.bgz.tbi").exists():
response = requests.get("https://storage.googleapis.com/sgkit-data/tutorial/1kg.vcf.bgz.tbi")
with open("1kg.vcf.bgz.tbi", "wb") as f:
f.write(response.content)
Importing data from VCF#
Next, convert the VCF file to Zarr using the vcf2zarr command in bio2zarr, stored on the local filesystem in a directory called 1kg.vcz.
%%bash
vcf2zarr explode --force 1kg.vcf.bgz 1kg.icf
# vcf2zarr mkschema 1kg.icf > 1kg.schema.json # then edit 1kg.schema.json by hand
vcf2zarr encode --force -s 1kg.schema.json 1kg.icf 1kg.vcz
Scan: 0%| | 0.00/1.00 [00:00<?, ?files/s]
[W::bcf_hdr_check_sanity] PL should be declared as Number=G
Scan: 0%| | 0.00/1.00 [00:00<?, ?files/s]
Scan: 0%| | 0.00/1.00 [00:00<?, ?files/s]
Scan: 100%|██████████| 1.00/1.00 [00:00<00:00, 4.02files/s]
Explode: 0%| | 0.00/10.9k [00:00<?, ?vars/s]
Explode: 0%| | 0.00/10.9k [00:00<?, ?vars/s]
Explode: 0%| | 0.00/10.9k [00:00<?, ?vars/s]
Explode: 2%|▏ | 226/10.9k [00:00<00:04, 2.22kvars/s]
Explode: 4%|▎ | 395/10.9k [00:00<00:05, 1.93kvars/s]
Explode: 6%|▌ | 631/10.9k [00:00<00:04, 2.07kvars/s]
Explode: 8%|▊ | 867/10.9k [00:00<00:04, 2.14kvars/s]
Explode: 10%|▉ | 1.05k/10.9k [00:01<00:14, 661vars/s]
Explode: 12%|█▏ | 1.28k/10.9k [00:01<00:12, 796vars/s]
Explode: 14%|█▍ | 1.52k/10.9k [00:01<00:10, 924vars/s]
Explode: 16%|█▌ | 1.75k/10.9k [00:01<00:08, 1.03kvars/s]
Explode: 18%|█▊ | 1.94k/10.9k [00:02<00:13, 657vars/s]
Explode: 20%|██ | 2.19k/10.9k [00:02<00:11, 750vars/s]
Explode: 22%|██▏ | 2.43k/10.9k [00:03<00:10, 839vars/s]
Explode: 24%|██▍ | 2.61k/10.9k [00:03<00:13, 622vars/s]
Explode: 26%|██▌ | 2.85k/10.9k [00:03<00:11, 697vars/s]
Explode: 28%|██▊ | 3.10k/10.9k [00:04<00:10, 774vars/s]
Explode: 30%|███ | 3.28k/10.9k [00:04<00:12, 617vars/s]
Explode: 32%|███▏ | 3.52k/10.9k [00:05<00:10, 685vars/s]
Explode: 35%|███▍ | 3.76k/10.9k [00:05<00:09, 756vars/s]
Explode: 36%|███▋ | 3.96k/10.9k [00:05<00:11, 616vars/s]
Explode: 38%|███▊ | 4.16k/10.9k [00:06<00:10, 665vars/s]
Explode: 40%|████ | 4.41k/10.9k [00:06<00:08, 732vars/s]
Explode: 42%|████▏ | 4.58k/10.9k [00:06<00:10, 597vars/s]
Explode: 44%|████▍ | 4.82k/10.9k [00:07<00:09, 657vars/s]
Explode: 46%|████▌ | 5.00k/10.9k [00:07<00:08, 697vars/s]
Explode: 48%|████▊ | 5.17k/10.9k [00:07<00:09, 602vars/s]
Explode: 50%|████▉ | 5.41k/10.9k [00:07<00:08, 663vars/s]
Explode: 51%|█████▏ | 5.58k/10.9k [00:08<00:09, 577vars/s]
Explode: 53%|█████▎ | 5.82k/10.9k [00:08<00:07, 636vars/s]
Explode: 55%|█████▍ | 5.98k/10.9k [00:09<00:08, 568vars/s]
Explode: 57%|█████▋ | 6.22k/10.9k [00:09<00:07, 626vars/s]
Explode: 59%|█████▉ | 6.45k/10.9k [00:09<00:06, 682vars/s]
Explode: 61%|██████ | 6.61k/10.9k [00:10<00:07, 586vars/s]
Explode: 63%|██████▎ | 6.86k/10.9k [00:10<00:06, 644vars/s]
Explode: 65%|██████▍ | 7.02k/10.9k [00:10<00:05, 678vars/s]
Explode: 66%|██████▌ | 7.19k/10.9k [00:11<00:06, 582vars/s]
Explode: 68%|██████▊ | 7.43k/10.9k [00:11<00:05, 640vars/s]
Explode: 70%|██████▉ | 7.59k/10.9k [00:11<00:04, 673vars/s]
Explode: 71%|███████▏ | 7.75k/10.9k [00:12<00:05, 577vars/s]
Explode: 73%|███████▎ | 7.92k/10.9k [00:12<00:04, 614vars/s]
Explode: 74%|███████▍ | 8.07k/10.9k [00:12<00:04, 565vars/s]
Explode: 76%|███████▌ | 8.26k/10.9k [00:12<00:04, 608vars/s]
Explode: 77%|███████▋ | 8.42k/10.9k [00:13<00:04, 560vars/s]
Explode: 79%|███████▉ | 8.60k/10.9k [00:13<00:03, 601vars/s]
Explode: 80%|████████ | 8.75k/10.9k [00:13<00:03, 552vars/s]
Explode: 83%|████████▎ | 8.99k/10.9k [00:14<00:03, 610vars/s]
Explode: 84%|████████▍ | 9.13k/10.9k [00:14<00:03, 553vars/s]
Explode: 86%|████████▌ | 9.33k/10.9k [00:14<00:02, 600vars/s]
Explode: 87%|████████▋ | 9.48k/10.9k [00:15<00:02, 547vars/s]
Explode: 89%|████████▊ | 9.63k/10.9k [00:15<00:02, 577vars/s]
Explode: 90%|████████▉ | 9.76k/10.9k [00:15<00:02, 537vars/s]
Explode: 92%|█████████▏| 9.97k/10.9k [00:15<00:01, 589vars/s]
Explode: 93%|█████████▎| 10.1k/10.9k [00:16<00:01, 541vars/s]
Explode: 95%|█████████▍| 10.3k/10.9k [00:16<00:01, 580vars/s]
Explode: 96%|█████████▌| 10.4k/10.9k [00:17<00:00, 530vars/s]
Explode: 97%|█████████▋| 10.5k/10.9k [00:17<00:00, 504vars/s]
Explode: 98%|█████████▊| 10.6k/10.9k [00:17<00:00, 478vars/s]
Explode: 100%|██████████| 10.9k/10.9k [00:17<00:00, 539vars/s]
Explode: 100%|██████████| 10.9k/10.9k [00:18<00:00, 601vars/s]
Encode: 0%| | 0.00/28.9M [00:00<?, ?B/s]
Encode: 0%| | 120k/28.9M [00:00<00:25, 1.12MB/s]
Encode: 6%|▋ | 1.86M/28.9M [00:00<00:02, 9.30MB/s]
Encode: 12%|█▏ | 3.60M/28.9M [00:00<00:02, 12.1MB/s]
Encode: 18%|█▊ | 5.33M/28.9M [00:00<00:01, 13.5MB/s]
Encode: 25%|██▍ | 7.09M/28.9M [00:00<00:01, 14.4MB/s]
Encode: 33%|███▎ | 9.59M/28.9M [00:00<00:01, 16.6MB/s]
Encode: 40%|███▉ | 11.5M/28.9M [00:01<00:02, 6.98MB/s]
Encode: 46%|████▌ | 13.2M/28.9M [00:01<00:02, 7.14MB/s]
Encode: 52%|█████▏ | 14.9M/28.9M [00:01<00:01, 7.32MB/s]
Encode: 57%|█████▋ | 16.5M/28.9M [00:02<00:01, 7.44MB/s]
Encode: 62%|██████▏ | 18.0M/28.9M [00:02<00:01, 7.88MB/s]
Encode: 67%|██████▋ | 19.4M/28.9M [00:02<00:01, 8.28MB/s]
Encode: 73%|███████▎ | 21.0M/28.9M [00:02<00:00, 8.83MB/s]
Encode: 79%|███████▊ | 22.7M/28.9M [00:02<00:00, 9.37MB/s]
Encode: 100%|██████████| 28.9M/28.9M [00:02<00:00, 11.1MB/s]
Finalise: 0%| | 0.00/32.0 [00:00<?, ?array/s]
Finalise: 100%|██████████| 32.0/32.0 [00:00<00:00, 3.07karray/s]
We used the vcf2zarr explode command to first convert the VCF to an “intermediate columnar format” (ICF), then the vcf2zarr encode command to convert the ICF to Zarr, which by convention is stored in a directory with a vcz extension.
Note that we specified a JSON schema file that was created with the vcf2zarr mkschema command (commented out above), then edited to drop some fields that are not needed for this tutorial (such as FORMAT/PL). It was also updated to change the call_AD field’s third dimension to be alleles, which was not set by vcf2zarr since the dataset we are using defines FORMAT/AD as . which means “unknown”, rather than R.
For more information about using vcf2zarr, see the tutorial in the bio2zarr documentation.
Now the data has been written as Zarr, all downstream operations on will be much faster. Note that sgkit uses an Xarray dataset to represent the VCF data, where Hail uses MatrixTable.
ds = sg.load_dataset("1kg.vcz")
Getting to know our data#
To start with we’ll look at some summary data from the dataset.
The simplest thing is to look at the dimensions and data variables in the Xarray dataset.
ds
<xarray.Dataset> Size: 29MB
Dimensions: (variants: 10879, samples: 284, alleles: 2,
ploidy: 2, contigs: 84, filters: 1,
region_index_values: 33, region_index_fields: 6)
Dimensions without coordinates: variants, samples, alleles, ploidy, contigs,
filters, region_index_values,
region_index_fields
Data variables: (12/38)
call_AD (variants, samples, alleles) int8 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_DP (variants, samples) int8 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
call_GQ (variants, samples) int8 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
call_genotype (variants, samples, ploidy) int8 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_genotype_mask (variants, samples, ploidy) bool 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_genotype_phased (variants, samples) bool 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
... ...
variant_id (variants) object 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_id_mask (variants) bool 11kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_length (variants) int8 11kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_position (variants) int32 44kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_quality (variants) float32 44kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_set (variants) object 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
Attributes: (3)- variants: 10879
- samples: 284
- alleles: 2
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- call_AD(variants, samples, alleles)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - sample_id(samples)objectdask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(1000, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(1000, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879, 1) (1000, 1) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
Next we’ll use display_genotypes to show the the first and last few variants and samples.
Note: sgkit does not store the contig names in an easily accessible form, so we compute a variable variant_contig_name in the same dataset storing them for later use, and set an index so we can see the variant name, position, and ID.
ds["variant_contig_name"] = ds.contig_id[ds.variant_contig]
ds2 = ds.set_index({"variants": ("variant_contig_name", "variant_position", "variant_id")})
sg.display_genotypes(ds2, max_variants=10, max_samples=5)
| samples | HG00096 | HG00099 | ... | NA21133 | NA21143 |
|---|---|---|---|---|---|
| variants | |||||
| (1, 904165, .) | 0/0 | 0/0 | ... | 0/0 | 0/0 |
| (1, 909917, .) | 0/0 | 0/0 | ... | 0/0 | 0/0 |
| (1, 986963, .) | 0/0 | 0/0 | ... | 0/0 | 0/0 |
| (1, 1563691, .) | ./. | 0/0 | ... | 0/0 | 0/0 |
| (1, 1707740, .) | 0/1 | 0/1 | ... | 0/1 | 0/0 |
| ... | ... | ... | ... | ... | ... |
| (X, 152660491, .) | ./. | 0/0 | ... | 1/1 | 0/0 |
| (X, 153031688, .) | 0/0 | 0/0 | ... | 0/0 | 0/0 |
| (X, 153674876, .) | 0/0 | 0/0 | ... | 0/0 | 0/0 |
| (X, 153706320, .) | ./. | 0/0 | ... | 0/0 | 0/0 |
| (X, 154087368, .) | 0/0 | 1/1 | ... | 1/1 | 1/1 |
10879 rows x 284 columns
We can show the alleles too.
Note: this needs work to make it easier to do
df_variant = ds[[v for v in ds.data_vars if v.startswith("variant_")]].to_dataframe()
df_variant.groupby(["variant_contig_name", "variant_position", "variant_id"]).agg({"variant_allele": lambda x: list(x)}).head(5)
| variant_allele | |||
|---|---|---|---|
| variant_contig_name | variant_position | variant_id | |
| 1 | 904165 | . | [G, A] |
| 909917 | . | [G, A] | |
| 986963 | . | [C, T] | |
| 1563691 | . | [T, G] | |
| 1707740 | . | [T, G] |
Show the first five sample IDs by referencing the dataset variable directly:
ds.sample_id[:5].values
array(['HG00096', 'HG00099', 'HG00105', 'HG00118', 'HG00129'],
dtype=object)
Adding column fields#
Xarray datasets can have any number of variables added to them, possibly loaded from different sources. Next we’ll take a text file (CSV) containing annotations, and use it to annotate the samples in the dataset.
First we load the annotation data using regular Pandas.
ANNOTATIONS_FILE = "https://storage.googleapis.com/sgkit-gwas-tutorial/1kg_annotations.txt"
df = pd.read_csv(ANNOTATIONS_FILE, sep="\t", index_col="Sample")
df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 3500 entries, HG00096 to NA21144
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Population 3500 non-null object
1 SuperPopulation 3500 non-null object
2 isFemale 3500 non-null bool
3 PurpleHair 3500 non-null bool
4 CaffeineConsumption 3500 non-null int64
dtypes: bool(2), int64(1), object(2)
memory usage: 116.2+ KB
df
| Population | SuperPopulation | isFemale | PurpleHair | CaffeineConsumption | |
|---|---|---|---|---|---|
| Sample | |||||
| HG00096 | GBR | EUR | False | False | 4 |
| HG00097 | GBR | EUR | True | True | 4 |
| HG00098 | GBR | EUR | False | False | 5 |
| HG00099 | GBR | EUR | True | False | 4 |
| HG00100 | GBR | EUR | True | False | 5 |
| ... | ... | ... | ... | ... | ... |
| NA21137 | GIH | SAS | True | False | 1 |
| NA21141 | GIH | SAS | True | True | 2 |
| NA21142 | GIH | SAS | True | True | 2 |
| NA21143 | GIH | SAS | True | True | 5 |
| NA21144 | GIH | SAS | True | False | 3 |
3500 rows × 5 columns
To join the annotation data with the genetic data, we convert it to Xarray, then do a join.
ds_annotations = pd.DataFrame.to_xarray(df).rename({"Sample":"samples"})
ds = ds.set_index({"samples": "sample_id"})
ds = ds.merge(ds_annotations, join="left")
ds = ds.reset_index("samples").reset_coords(drop=True)
ds
<xarray.Dataset> Size: 29MB
Dimensions: (samples: 284, variants: 10879, alleles: 2,
ploidy: 2, contigs: 84, filters: 1,
region_index_values: 33, region_index_fields: 6)
Dimensions without coordinates: samples, variants, alleles, ploidy, contigs,
filters, region_index_values,
region_index_fields
Data variables: (12/43)
call_AD (variants, samples, alleles) int8 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_DP (variants, samples) int8 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
call_GQ (variants, samples) int8 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
call_genotype (variants, samples, ploidy) int8 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_genotype_mask (variants, samples, ploidy) bool 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_genotype_phased (variants, samples) bool 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
... ...
variant_contig_name (variants) object 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
Population (samples) object 2kB 'GBR' 'GBR' ... 'GIH' 'GIH'
SuperPopulation (samples) object 2kB 'EUR' 'EUR' ... 'SAS' 'SAS'
isFemale (samples) bool 284B False True False ... False True
PurpleHair (samples) bool 284B False False False ... True True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 6 2 4 ... 6 4 6 4 6 5 5
Attributes: (3)- samples: 284
- variants: 10879
- alleles: 2
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- call_AD(variants, samples, alleles)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(1000, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(1000, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879, 1) (1000, 1) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'ACB', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AFR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, False, True, False, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, False, True, True, True, False, True, True, False, False, False, True, False, True, False, False, True, True, True, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, False, True, False, True, True, True, False, False, True, False, False, True, False, True, False, True, False, False, True, True, False, False, False, True, False, True, True, True, False, True, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, True, False, False, False, True, False, True, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, True, False, True, True, False, True, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, False, True, True, False, True, True, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, False, False, True, True, False, True, True, False, True, True, False, False, True, False, True, True, True, False, False, False, False, False, False, True, False, False, False, True, False, False, True, False, True, True, False, False, False, False, False, True, False, False, True, False, True, True, True, False, False, False, False, True, False, False, True, True, False, True, True, False, False, False, False, True, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 4 2 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 4, 2, 1, 2, 0, 5, 4, 5, 4, 3, 6, 5, 5, 7, 5, 5, 7, 5, 1, 5, 5, 5, 4, 4, 5, 5, 5, 6, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 4, 7, 5, 4, 2, 6, 7, 6, 5, 3, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 3, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 4, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 5, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 6, 5, 4, 3, 8, 2, 4, 4, 6, 8, 4, 3, 4, 3, 2, 5, 6, 6, 4, 3, 5, 7, 4, 2, 5, 5, 6, 3, 2, 4, 4, 6, 5, 6, 5, 7, 2, 4, 2, 1, 5, 3, 5, 3, 5, 2, 4, 9, 6, 4, 3, 4, 4, 6, 6, 7, 6, 6, 3, 4, 3, 6, 6, 3, 4, 4, 2, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5])
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
Query functions#
We can look at some statistics of the data by converting the relevant dataset variable to a Pandas series, then using its built-in summary functions. Annotation data is usually small enough to load into memory, which is why it’s OK using Pandas here.
Here’s the population distribution by continent:
ds_annotations.SuperPopulation.to_series().value_counts()
SuperPopulation
AFR 1018
EUR 669
SAS 661
EAS 617
AMR 535
Name: count, dtype: int64
The distribution of the CaffeineConsumption variable:
ds_annotations.CaffeineConsumption.to_series().describe()
count 3500.000000
mean 3.983714
std 1.702349
min -1.000000
25% 3.000000
50% 4.000000
75% 5.000000
max 10.000000
Name: CaffeineConsumption, dtype: float64
There are far fewer samples in our dataset than the full 1000 genomes dataset, as we can see from the following queries.
len(ds_annotations.samples)
3500
len(ds.samples)
284
ds.SuperPopulation.to_series().value_counts()
SuperPopulation
AFR 76
EAS 72
SAS 55
EUR 47
AMR 34
Name: count, dtype: int64
ds.CaffeineConsumption.to_series().describe()
count 284.000000
mean 4.415493
std 1.580549
min 0.000000
25% 3.000000
50% 4.000000
75% 5.000000
max 9.000000
Name: CaffeineConsumption, dtype: float64
Here’s an example of doing an ad hoc query to uncover a biological insight from the data: calculate the counts of each of the 12 possible unique SNPs (4 choices for the reference base * 3 choices for the alternate base).
df_variant.groupby(["variant_contig_name", "variant_position", "variant_id"])["variant_allele"].apply(tuple).value_counts()
variant_allele
(C, T) 2418
(G, A) 2367
(A, G) 1929
(T, C) 1864
(C, A) 494
(G, T) 477
(T, G) 466
(A, C) 451
(C, G) 150
(G, C) 111
(T, A) 77
(A, T) 75
Name: count, dtype: int64
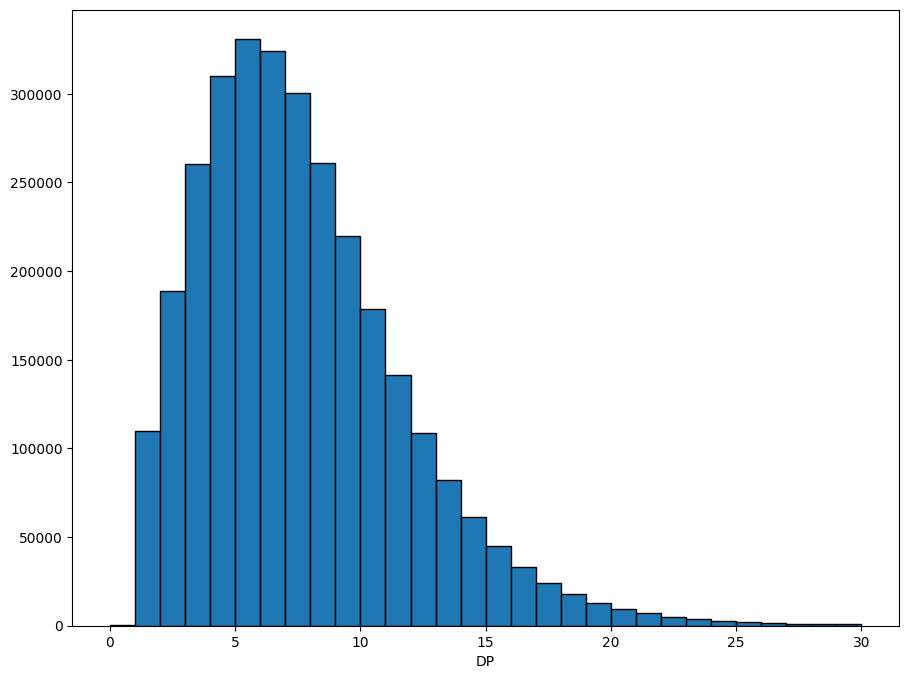
Often we want to plot the data, to get a feel for how it’s distributed. Xarray has some convenience functions for plotting, which we use here to show the distribution of the DP field.
dp = ds.call_DP.where(ds.call_DP >= 0) # filter out missing
dp.attrs["long_name"] = "DP"
xr.plot.hist(dp, range=(0, 30), bins=30, size=8, edgecolor="black");
Quality control#
QC is the process of filtering out poor quality data before running an analysis. This is usually an iterative process.
The sample_stats function in sgkit computes a collection of useful metrics for each sample and stores them in new variables. (The Hail equivalent is sample_qc.)
Here’s the dataset before running sample_stats.
ds
<xarray.Dataset> Size: 29MB
Dimensions: (samples: 284, variants: 10879, alleles: 2,
ploidy: 2, contigs: 84, filters: 1,
region_index_values: 33, region_index_fields: 6)
Dimensions without coordinates: samples, variants, alleles, ploidy, contigs,
filters, region_index_values,
region_index_fields
Data variables: (12/43)
call_AD (variants, samples, alleles) int8 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_DP (variants, samples) int8 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
call_GQ (variants, samples) int8 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
call_genotype (variants, samples, ploidy) int8 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_genotype_mask (variants, samples, ploidy) bool 6MB dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
call_genotype_phased (variants, samples) bool 3MB dask.array<chunksize=(1000, 284), meta=np.ndarray>
... ...
variant_contig_name (variants) object 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
Population (samples) object 2kB 'GBR' 'GBR' ... 'GIH' 'GIH'
SuperPopulation (samples) object 2kB 'EUR' 'EUR' ... 'SAS' 'SAS'
isFemale (samples) bool 284B False True False ... False True
PurpleHair (samples) bool 284B False False False ... True True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 6 2 4 ... 6 4 6 4 6 5 5
Attributes: (3)- samples: 284
- variants: 10879
- alleles: 2
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- call_AD(variants, samples, alleles)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(1000, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(1000, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879, 1) (1000, 1) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'ACB', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AFR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, False, True, False, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, False, True, True, True, False, True, True, False, False, False, True, False, True, False, False, True, True, True, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, False, True, False, True, True, True, False, False, True, False, False, True, False, True, False, True, False, False, True, True, False, False, False, True, False, True, True, True, False, True, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, True, False, False, False, True, False, True, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, True, False, True, True, False, True, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, False, True, True, False, True, True, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, False, False, True, True, False, True, True, False, True, True, False, False, True, False, True, True, True, False, False, False, False, False, False, True, False, False, False, True, False, False, True, False, True, True, False, False, False, False, False, True, False, False, True, False, True, True, True, False, False, False, False, True, False, False, True, True, False, True, True, False, False, False, False, True, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 4 2 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 4, 2, 1, 2, 0, 5, 4, 5, 4, 3, 6, 5, 5, 7, 5, 5, 7, 5, 1, 5, 5, 5, 4, 4, 5, 5, 5, 6, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 4, 7, 5, 4, 2, 6, 7, 6, 5, 3, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 3, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 4, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 5, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 6, 5, 4, 3, 8, 2, 4, 4, 6, 8, 4, 3, 4, 3, 2, 5, 6, 6, 4, 3, 5, 7, 4, 2, 5, 5, 6, 3, 2, 4, 4, 6, 5, 6, 5, 7, 2, 4, 2, 1, 5, 3, 5, 3, 5, 2, 4, 9, 6, 4, 3, 4, 4, 6, 6, 7, 6, 6, 3, 4, 3, 6, 6, 3, 4, 4, 2, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5])
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
We can see the new variables (with names beginning sample_) after we run sample_stats:
ds = sg.sample_stats(ds)
ds
<xarray.Dataset> Size: 29MB
Dimensions: (samples: 284, variants: 10879, alleles: 2,
ploidy: 2, contigs: 84, filters: 1,
region_index_values: 33, region_index_fields: 6)
Dimensions without coordinates: samples, variants, alleles, ploidy, contigs,
filters, region_index_values,
region_index_fields
Data variables: (12/49)
sample_n_called (samples) int64 2kB dask.array<chunksize=(284,), meta=np.ndarray>
sample_call_rate (samples) float64 2kB dask.array<chunksize=(284,), meta=np.ndarray>
sample_n_het (samples) int64 2kB dask.array<chunksize=(284,), meta=np.ndarray>
sample_n_hom_ref (samples) int64 2kB dask.array<chunksize=(284,), meta=np.ndarray>
sample_n_hom_alt (samples) int64 2kB dask.array<chunksize=(284,), meta=np.ndarray>
sample_n_non_ref (samples) int64 2kB dask.array<chunksize=(284,), meta=np.ndarray>
... ...
variant_contig_name (variants) object 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
Population (samples) object 2kB 'GBR' 'GBR' ... 'GIH' 'GIH'
SuperPopulation (samples) object 2kB 'EUR' 'EUR' ... 'SAS' 'SAS'
isFemale (samples) bool 284B False True False ... False True
PurpleHair (samples) bool 284B False False False ... True True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 6 2 4 ... 6 4 6 4 6 5 5
Attributes: (3)- samples: 284
- variants: 10879
- alleles: 2
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- sample_n_called(samples)int64dask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_call_rate(samples)float64dask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 11 graph layers Data type float64 numpy.ndarray - sample_n_het(samples)int64dask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 8 graph layers Data type int64 numpy.ndarray - sample_n_hom_ref(samples)int64dask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 8 graph layers Data type int64 numpy.ndarray - sample_n_hom_alt(samples)int64dask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 8 graph layers Data type int64 numpy.ndarray - sample_n_non_ref(samples)int64dask.array<chunksize=(284,), meta=np.ndarray>
Array Chunk Bytes 2.22 kiB 2.22 kiB Shape (284,) (284,) Dask graph 1 chunks in 11 graph layers Data type int64 numpy.ndarray - call_AD(variants, samples, alleles)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(1000, 284, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.89 MiB 554.69 kiB Shape (10879, 284, 2) (1000, 284, 2) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(1000, 284), meta=np.ndarray>
- description :
Array Chunk Bytes 2.95 MiB 277.34 kiB Shape (10879, 284) (1000, 284) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(1000, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(1000, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879, 1) (1000, 1) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'ACB', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AFR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, False, True, False, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, False, True, True, True, False, True, True, False, False, False, True, False, True, False, False, True, True, True, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, False, True, False, True, True, True, False, False, True, False, False, True, False, True, False, True, False, False, True, True, False, False, False, True, False, True, True, True, False, True, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, True, False, False, False, True, False, True, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, True, False, True, True, False, True, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, False, True, True, False, True, True, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, False, False, True, True, False, True, True, False, True, True, False, False, True, False, True, True, True, False, False, False, False, False, False, True, False, False, False, True, False, False, True, False, True, True, False, False, False, False, False, True, False, False, True, False, True, True, True, False, False, False, False, True, False, False, True, True, False, True, True, False, False, False, False, True, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 4 2 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 4, 2, 1, 2, 0, 5, 4, 5, 4, 3, 6, 5, 5, 7, 5, 5, 7, 5, 1, 5, 5, 5, 4, 4, 5, 5, 5, 6, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 4, 7, 5, 4, 2, 6, 7, 6, 5, 3, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 3, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 4, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 5, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 6, 5, 4, 3, 8, 2, 4, 4, 6, 8, 4, 3, 4, 3, 2, 5, 6, 6, 4, 3, 5, 7, 4, 2, 5, 5, 6, 3, 2, 4, 4, 6, 5, 6, 5, 7, 2, 4, 2, 1, 5, 3, 5, 3, 5, 2, 4, 9, 6, 4, 3, 4, 4, 6, 6, 7, 6, 6, 3, 4, 3, 6, 6, 3, 4, 4, 2, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5])
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
We can plot the metrics next.
ds.sample_call_rate.attrs["long_name"] = "Sample call rate"
xr.plot.hist(ds.sample_call_rate, range=(.88,1), bins=50, size=8, edgecolor="black");
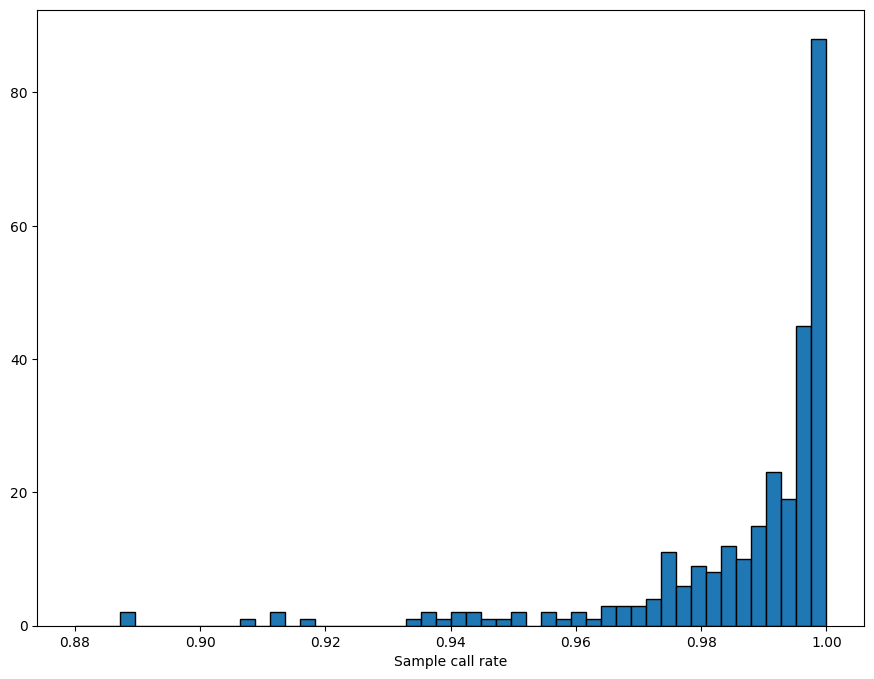
gq = ds.call_GQ.where(ds.call_GQ >= 0) # filter out missing
sample_gq_mean = gq.mean(dim="variants")
sample_gq_mean.attrs["long_name"] = "Mean Sample GQ"
xr.plot.hist(sample_gq_mean, range=(10,70), bins=60, size=8, edgecolor="black");
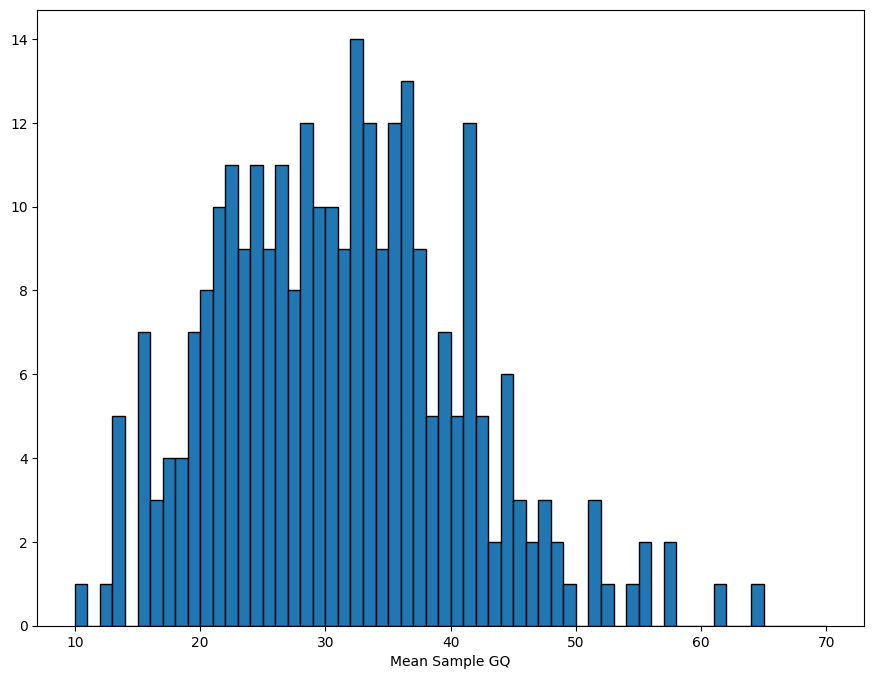
dp = ds.call_DP.where(ds.call_DP >= 0) # filter out missing
sample_dp_mean = dp.mean(dim="variants")
sample_dp_mean.attrs["long_name"] = "Mean Sample DP"
ds["sample_dp_mean"] = sample_dp_mean # add new data array to dataset
# Following does not work with recent versions of xarray, see https://github.com/sgkit-dev/sgkit/issues/934
#ds.plot.scatter(x="sample_dp_mean", y="sample_call_rate", size=8, s=10);
The following removes outliers using an arbitrary cutoff.
ds = ds.sel(samples=((ds.sample_dp_mean >= 4) & (ds.sample_call_rate >= 0.97)).compute())
print(f"After filter, {len(ds.samples)}/284 samples remain.")
After filter, 250/284 samples remain.
Genotype QC is more complicated. First we calculate a variable ab, which is the fraction of reads that were the alternate allele.
# fill rows with nan where no alternate alleles were read or where sum of reads is 0
ad1 = ds.call_AD.sel(dict(alleles=1)).pipe(lambda v: v.where(v >= 0))
adsum = ds.call_AD.sum(dim="alleles").pipe(lambda v: v.where(v != 0))
# compute alternate allele read fraction
ab = ad1 / adsum
Then we can use the ab variable in the filter condition, to filter homozygous reference calls with >10% alternate reads, homozygous alternate calls with >10% reference reads, or heterozygote calls without a ref/alt balance near 50%.
GT = ds.call_genotype
hom_ref = (GT == 0).all(dim="ploidy")
het = GT[..., 0] != GT[..., 1]
hom_alt = ((GT > 0) & (GT[..., 0] == GT)).all(dim="ploidy")
filter_condition_ab = ((hom_ref & (ab <= 0.1)) |
(het & (ab >= 0.25) & (ab <= 0.75)) |
(hom_alt & (ab >= 0.9)))
filter_mask = xr.where(ds.call_genotype_mask, True, filter_condition_ab)
fraction_filtered = GT.where(~filter_mask).count().values / GT.size
print(f"Filtering {fraction_filtered * 100:.2f}% entries out of downstream analysis.")
Filtering 3.65% entries out of downstream analysis.
Note: genotype QC is filtering out slightly different numbers of entries compared to the Hail tutorial.
Variant QC is similar. This time we use the variant_stats function, but we won’t do any filtering on these variables.
ds = sg.variant_stats(ds)
ds
<xarray.Dataset> Size: 27MB
Dimensions: (variants: 10879, alleles: 2, samples: 250,
ploidy: 2, contigs: 84, filters: 1,
region_index_values: 33, region_index_fields: 6)
Dimensions without coordinates: variants, alleles, samples, ploidy, contigs,
filters, region_index_values,
region_index_fields
Data variables: (12/59)
variant_n_called (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_call_rate (variants) float64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_n_het (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_n_hom_ref (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_n_hom_alt (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_n_non_ref (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
... ...
Population (samples) object 2kB 'GBR' 'GBR' ... 'GIH' 'GIH'
SuperPopulation (samples) object 2kB 'EUR' 'EUR' ... 'SAS' 'SAS'
isFemale (samples) bool 250B False True ... False True
PurpleHair (samples) bool 250B False False ... True True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 6 2 2 ... 4 6 4 6 5 5
sample_dp_mean (samples) float32 1kB dask.array<chunksize=(250,), meta=np.ndarray>
Attributes: (3)- variants: 10879
- alleles: 2
- samples: 250
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- variant_n_called(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_call_rate(variants)float64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 10 graph layers Data type float64 numpy.ndarray - variant_n_het(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 7 graph layers Data type int64 numpy.ndarray - variant_n_hom_ref(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 7 graph layers Data type int64 numpy.ndarray - variant_n_hom_alt(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 7 graph layers Data type int64 numpy.ndarray - variant_n_non_ref(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 10 graph layers Data type int64 numpy.ndarray - variant_allele_count(variants, alleles)uint64dask.array<chunksize=(1000, 2), meta=np.ndarray>
- comment :
- Variant allele counts. With shape (variants, alleles) and values corresponding to the number of non-missing occurrences of each allele.
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 5 graph layers Data type uint64 numpy.ndarray - variant_allele_total(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_allele_frequency(variants, alleles)float64dask.array<chunksize=(1000, 2), meta=np.ndarray>
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 13 graph layers Data type float64 numpy.ndarray - sample_n_called(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 10 graph layers Data type int64 numpy.ndarray - sample_call_rate(samples)float64dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Sample call rate
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type float64 numpy.ndarray - sample_n_het(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_alt(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_non_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type int64 numpy.ndarray - call_AD(variants, samples, alleles)int8dask.array<chunksize=(1000, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.19 MiB 488.28 kiB Shape (10879, 250, 2) (1000, 250, 2) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(1000, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.59 MiB 244.14 kiB Shape (10879, 250) (1000, 250) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(1000, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.59 MiB 244.14 kiB Shape (10879, 250) (1000, 250) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(1000, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.19 MiB 488.28 kiB Shape (10879, 250, 2) (1000, 250, 2) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(1000, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.19 MiB 488.28 kiB Shape (10879, 250, 2) (1000, 250, 2) Dask graph 11 chunks in 3 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(1000, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.59 MiB 244.14 kiB Shape (10879, 250) (1000, 250) Dask graph 11 chunks in 3 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(1000, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(1000, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879, 1) (1000, 1) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, True, True, True, True, False, False, False, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, True, False, True, False, False, True, True, False, False, True, True, True, False, False, False, False, True, False, False, True, True, True, False, True, False, True, False, True, True, True, False, True, False, True, False, True, False, False, True, False, False, True, False, True, True, True, False, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, False, False, True, True, False, False, True, True, True, True, True, True, True, False, True, True, False, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, True, True, False, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, True, False, False, False, True, False, False, True, False, False, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 2 5 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 2, 5, 5, 4, 6, 5, 7, 5, 7, 1, 5, 5, 4, 4, 5, 5, 5, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 7, 5, 2, 6, 7, 6, 5, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 4, 3, 8, 4, 6, 8, 3, 4, 2, 5, 6, 6, 4, 3, 5, 7, 4, 5, 5, 6, 3, 2, 4, 5, 6, 7, 2, 4, 2, 1, 5, 3, 4, 9, 6, 4, 3, 4, 4, 6, 7, 6, 3, 4, 3, 6, 6, 3, 4, 4, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5]) - sample_dp_mean(samples)float32dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Mean Sample DP
Array Chunk Bytes 0.98 kiB 0.98 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type float32 numpy.ndarray
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
Let’s do a GWAS!#
First we need to restrict to common variants (1% cutoff), and not far from Hardy-Weinberg equilibrium.
ds = sg.hardy_weinberg_test(ds)
(The warning is telling us that some variables are being regenerated, and can be safely ignored.)
ds
<xarray.Dataset> Size: 27MB
Dimensions: (variants: 10879, genotypes: 3, alleles: 2,
samples: 250, ploidy: 2, contigs: 84,
filters: 1, region_index_values: 33,
region_index_fields: 6)
Coordinates:
* genotypes (genotypes) <U3 36B '0/0' '0/1' '1/1'
Dimensions without coordinates: variants, alleles, samples, ploidy, contigs,
filters, region_index_values,
region_index_fields
Data variables: (12/62)
variant_hwe_p_value (variants) float64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_genotype_count (variants, genotypes) uint64 261kB dask.array<chunksize=(1000, 3), meta=np.ndarray>
genotype_id (genotypes) <U3 36B dask.array<chunksize=(3,), meta=np.ndarray>
variant_n_called (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_call_rate (variants) float64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
variant_n_het (variants) int64 87kB dask.array<chunksize=(1000,), meta=np.ndarray>
... ...
Population (samples) object 2kB 'GBR' 'GBR' ... 'GIH' 'GIH'
SuperPopulation (samples) object 2kB 'EUR' 'EUR' ... 'SAS' 'SAS'
isFemale (samples) bool 250B False True ... False True
PurpleHair (samples) bool 250B False False ... True True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 6 2 2 ... 4 6 4 6 5 5
sample_dp_mean (samples) float32 1kB dask.array<chunksize=(250,), meta=np.ndarray>
Attributes: (3)- variants: 10879
- genotypes: 3
- alleles: 2
- samples: 250
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- genotypes(genotypes)<U3'0/0' '0/1' '1/1'
array(['0/0', '0/1', '1/1'], dtype='<U3')
- variant_hwe_p_value(variants)float64dask.array<chunksize=(1000,), meta=np.ndarray>
- comment :
- P values from HWE test for each variant as float in [0, 1].
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type float64 numpy.ndarray - variant_genotype_count(variants, genotypes)uint64dask.array<chunksize=(1000, 3), meta=np.ndarray>
- comment :
- The number of observations for each possible genotype at each variant. Counts are sorted following the ordering defined in the VCF specification. - For biallelic, diploid genotypes the ordering is ``00``, ``01``, ``11`` (homozygous reference, heterozygous, homozygous alternate). - For triallelic, diploid genotypes the ordering is ``00``, ``01``, ``11``, ``02``, ``12``, ``22`` - For triallelic, triploid genotypes the ordering is ``000``, ``001``, ``011``, ``111``, ``002``, ``012``, ``112``, ``022``, ``122``, ``222``
Array Chunk Bytes 254.98 kiB 23.44 kiB Shape (10879, 3) (1000, 3) Dask graph 11 chunks in 4 graph layers Data type uint64 numpy.ndarray - genotype_id(genotypes)<U3dask.array<chunksize=(3,), meta=np.ndarray>
- comment :
- VCF style genotype strings for all possible genotypes given the size of the ploidy and alleles dimensions. The ordering of genotype strings follows the ordering outlined in the VCF specification for arrays of size "G".
Array Chunk Bytes 36 B 36 B Shape (3,) (3,) Dask graph 1 chunks in 4 graph layers Data type - variant_n_called(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_call_rate(variants)float64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 10 graph layers Data type float64 numpy.ndarray - variant_n_het(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 7 graph layers Data type int64 numpy.ndarray - variant_n_hom_ref(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 7 graph layers Data type int64 numpy.ndarray - variant_n_hom_alt(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 7 graph layers Data type int64 numpy.ndarray - variant_n_non_ref(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 10 graph layers Data type int64 numpy.ndarray - variant_allele_count(variants, alleles)uint64dask.array<chunksize=(1000, 2), meta=np.ndarray>
- comment :
- Variant allele counts. With shape (variants, alleles) and values corresponding to the number of non-missing occurrences of each allele.
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 5 graph layers Data type uint64 numpy.ndarray - variant_allele_total(variants)int64dask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_allele_frequency(variants, alleles)float64dask.array<chunksize=(1000, 2), meta=np.ndarray>
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 13 graph layers Data type float64 numpy.ndarray - sample_n_called(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 10 graph layers Data type int64 numpy.ndarray - sample_call_rate(samples)float64dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Sample call rate
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type float64 numpy.ndarray - sample_n_het(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_alt(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_non_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type int64 numpy.ndarray - call_AD(variants, samples, alleles)int8dask.array<chunksize=(1000, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.19 MiB 488.28 kiB Shape (10879, 250, 2) (1000, 250, 2) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(1000, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.59 MiB 244.14 kiB Shape (10879, 250) (1000, 250) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(1000, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.59 MiB 244.14 kiB Shape (10879, 250) (1000, 250) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(1000, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.19 MiB 488.28 kiB Shape (10879, 250, 2) (1000, 250, 2) Dask graph 11 chunks in 3 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(1000, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 5.19 MiB 488.28 kiB Shape (10879, 250, 2) (1000, 250, 2) Dask graph 11 chunks in 3 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(1000, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.59 MiB 244.14 kiB Shape (10879, 250) (1000, 250) Dask graph 11 chunks in 3 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 21.25 kiB 1.95 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(1000, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 169.98 kiB 15.62 kiB Shape (10879, 2) (1000, 2) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(1000, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879, 1) (1000, 1) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 10.62 kiB 0.98 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(1000,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 42.50 kiB 3.91 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
- description :
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 2 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(1000,), meta=np.ndarray>
Array Chunk Bytes 84.99 kiB 7.81 kiB Shape (10879,) (1000,) Dask graph 11 chunks in 8 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, True, True, True, True, False, False, False, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, True, False, True, False, False, True, True, False, False, True, True, True, False, False, False, False, True, False, False, True, True, True, False, True, False, True, False, True, True, True, False, True, False, True, False, True, False, False, True, False, False, True, False, True, True, True, False, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, False, False, True, True, False, False, True, True, True, True, True, True, True, False, True, True, False, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, True, True, False, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, True, False, False, False, True, False, False, True, False, False, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 2 5 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 2, 5, 5, 4, 6, 5, 7, 5, 7, 1, 5, 5, 4, 4, 5, 5, 5, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 7, 5, 2, 6, 7, 6, 5, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 4, 3, 8, 4, 6, 8, 3, 4, 2, 5, 6, 6, 4, 3, 5, 7, 4, 5, 5, 6, 3, 2, 4, 5, 6, 7, 2, 4, 2, 1, 5, 3, 4, 9, 6, 4, 3, 4, 4, 6, 7, 6, 3, 4, 3, 6, 6, 3, 4, 4, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5]) - sample_dp_mean(samples)float32dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Mean Sample DP
Array Chunk Bytes 0.98 kiB 0.98 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type float32 numpy.ndarray
- genotypesPandasIndex
PandasIndex(Index(['0/0', '0/1', '1/1'], dtype='object', name='genotypes'))
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
ds = ds.sel(variants=((ds.variant_allele_frequency[:,1] > 0.01) & (ds.variant_hwe_p_value > 1e-6)).compute())
Note: again, the number of variants is different to the Hail tutorial, but the final results work out to be very similar.
print(f"Samples: {len(ds.samples)} Variants: {len(ds.variants)}")
Samples: 250 Variants: 8394
Run a linear regression of dosage (number of alt alleles) against the CaffeineConsumption trait.
ds["call_dosage"] = ds.call_genotype.sum(dim="ploidy")
ds_lr = sg.gwas_linear_regression(ds, dosage="call_dosage",
add_intercept=True, covariates=[], traits=["CaffeineConsumption"])
ds_lr
<xarray.Dataset> Size: 38MB
Dimensions: (variants: 8394, traits: 1, genotypes: 3,
alleles: 2, samples: 250, ploidy: 2,
contigs: 84, filters: 1,
region_index_values: 33, region_index_fields: 6)
Coordinates:
* genotypes (genotypes) <U3 36B '0/0' '0/1' '1/1'
Dimensions without coordinates: variants, traits, alleles, samples, ploidy,
contigs, filters, region_index_values,
region_index_fields
Data variables: (12/66)
variant_linreg_beta (variants, traits) float64 67kB dask.array<chunksize=(989, 1), meta=np.ndarray>
variant_linreg_t_value (variants, traits) float64 67kB dask.array<chunksize=(989, 1), meta=np.ndarray>
variant_linreg_p_value (variants, traits) float64 67kB dask.array<chunksize=(989, 1), meta=np.ndarray>
variant_hwe_p_value (variants) float64 67kB dask.array<chunksize=(989,), meta=np.ndarray>
variant_genotype_count (variants, genotypes) uint64 201kB dask.array<chunksize=(989, 3), meta=np.ndarray>
genotype_id (genotypes) <U3 36B dask.array<chunksize=(3,), meta=np.ndarray>
... ...
SuperPopulation (samples) object 2kB 'EUR' 'EUR' ... 'SAS' 'SAS'
isFemale (samples) bool 250B False True ... False True
PurpleHair (samples) bool 250B False False ... True True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 6 2 2 ... 4 6 4 6 5 5
sample_dp_mean (samples) float32 1kB dask.array<chunksize=(250,), meta=np.ndarray>
call_dosage (variants, samples) int64 17MB dask.array<chunksize=(989, 250), meta=np.ndarray>
Attributes: (3)- variants: 8394
- traits: 1
- genotypes: 3
- alleles: 2
- samples: 250
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- genotypes(genotypes)<U3'0/0' '0/1' '1/1'
array(['0/0', '0/1', '1/1'], dtype='<U3')
- variant_linreg_beta(variants, traits)float64dask.array<chunksize=(989, 1), meta=np.ndarray>
- comment :
- Beta values associated with each variant and trait.
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394, 1) (989, 1) Dask graph 9 chunks in 33 graph layers Data type float64 numpy.ndarray - variant_linreg_t_value(variants, traits)float64dask.array<chunksize=(989, 1), meta=np.ndarray>
- comment :
- T statistics for each beta.
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394, 1) (989, 1) Dask graph 9 chunks in 45 graph layers Data type float64 numpy.ndarray - variant_linreg_p_value(variants, traits)float64dask.array<chunksize=(989, 1), meta=np.ndarray>
- comment :
- P values as float in [0, 1].
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394, 1) (989, 1) Dask graph 9 chunks in 47 graph layers Data type float64 numpy.ndarray - variant_hwe_p_value(variants)float64dask.array<chunksize=(989,), meta=np.ndarray>
- comment :
- P values from HWE test for each variant as float in [0, 1].
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 9 graph layers Data type float64 numpy.ndarray - variant_genotype_count(variants, genotypes)uint64dask.array<chunksize=(989, 3), meta=np.ndarray>
- comment :
- The number of observations for each possible genotype at each variant. Counts are sorted following the ordering defined in the VCF specification. - For biallelic, diploid genotypes the ordering is ``00``, ``01``, ``11`` (homozygous reference, heterozygous, homozygous alternate). - For triallelic, diploid genotypes the ordering is ``00``, ``01``, ``11``, ``02``, ``12``, ``22`` - For triallelic, triploid genotypes the ordering is ``000``, ``001``, ``011``, ``111``, ``002``, ``012``, ``112``, ``022``, ``122``, ``222``
Array Chunk Bytes 196.73 kiB 23.18 kiB Shape (8394, 3) (989, 3) Dask graph 9 chunks in 5 graph layers Data type uint64 numpy.ndarray - genotype_id(genotypes)<U3dask.array<chunksize=(3,), meta=np.ndarray>
- comment :
- VCF style genotype strings for all possible genotypes given the size of the ploidy and alleles dimensions. The ordering of genotype strings follows the ordering outlined in the VCF specification for arrays of size "G".
Array Chunk Bytes 36 B 36 B Shape (3,) (3,) Dask graph 1 chunks in 4 graph layers Data type - variant_n_called(variants)int64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 9 graph layers Data type int64 numpy.ndarray - variant_call_rate(variants)float64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 11 graph layers Data type float64 numpy.ndarray - variant_n_het(variants)int64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_n_hom_ref(variants)int64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_n_hom_alt(variants)int64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 8 graph layers Data type int64 numpy.ndarray - variant_n_non_ref(variants)int64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 11 graph layers Data type int64 numpy.ndarray - variant_allele_count(variants, alleles)uint64dask.array<chunksize=(989, 2), meta=np.ndarray>
- comment :
- Variant allele counts. With shape (variants, alleles) and values corresponding to the number of non-missing occurrences of each allele.
Array Chunk Bytes 131.16 kiB 15.45 kiB Shape (8394, 2) (989, 2) Dask graph 9 chunks in 6 graph layers Data type uint64 numpy.ndarray - variant_allele_total(variants)int64dask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 9 graph layers Data type int64 numpy.ndarray - variant_allele_frequency(variants, alleles)float64dask.array<chunksize=(989, 2), meta=np.ndarray>
Array Chunk Bytes 131.16 kiB 15.45 kiB Shape (8394, 2) (989, 2) Dask graph 9 chunks in 14 graph layers Data type float64 numpy.ndarray - sample_n_called(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 10 graph layers Data type int64 numpy.ndarray - sample_call_rate(samples)float64dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Sample call rate
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type float64 numpy.ndarray - sample_n_het(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_alt(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_non_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type int64 numpy.ndarray - call_AD(variants, samples, alleles)int8dask.array<chunksize=(989, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 4.00 MiB 482.91 kiB Shape (8394, 250, 2) (989, 250, 2) Dask graph 9 chunks in 4 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(989, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.00 MiB 241.46 kiB Shape (8394, 250) (989, 250) Dask graph 9 chunks in 4 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(989, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.00 MiB 241.46 kiB Shape (8394, 250) (989, 250) Dask graph 9 chunks in 4 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(989, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 4.00 MiB 482.91 kiB Shape (8394, 250, 2) (989, 250, 2) Dask graph 9 chunks in 4 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(989, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 4.00 MiB 482.91 kiB Shape (8394, 250, 2) (989, 250, 2) Dask graph 9 chunks in 4 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(989, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 2.00 MiB 241.46 kiB Shape (8394, 250) (989, 250) Dask graph 9 chunks in 4 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 16.39 kiB 1.93 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 16.39 kiB 1.93 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 8.20 kiB 0.97 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 16.39 kiB 1.93 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 8.20 kiB 0.97 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(989, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 131.16 kiB 15.45 kiB Shape (8394, 2) (989, 2) Dask graph 9 chunks in 3 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(989,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 8.20 kiB 0.97 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(989, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 8.20 kiB 0.97 kiB Shape (8394, 1) (989, 1) Dask graph 9 chunks in 3 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(989,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 8.20 kiB 0.97 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(989,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 8.20 kiB 0.97 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(989,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 32.79 kiB 3.86 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(989,), meta=np.ndarray>
- description :
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 3 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(989,), meta=np.ndarray>
Array Chunk Bytes 65.58 kiB 7.73 kiB Shape (8394,) (989,) Dask graph 9 chunks in 9 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, True, True, True, True, False, False, False, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, True, False, True, False, False, True, True, False, False, True, True, True, False, False, False, False, True, False, False, True, True, True, False, True, False, True, False, True, True, True, False, True, False, True, False, True, False, False, True, False, False, True, False, True, True, True, False, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, False, False, True, True, False, False, True, True, True, True, True, True, True, False, True, True, False, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, True, True, False, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, True, False, False, False, True, False, False, True, False, False, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 2 5 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 2, 5, 5, 4, 6, 5, 7, 5, 7, 1, 5, 5, 4, 4, 5, 5, 5, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 7, 5, 2, 6, 7, 6, 5, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 4, 3, 8, 4, 6, 8, 3, 4, 2, 5, 6, 6, 4, 3, 5, 7, 4, 5, 5, 6, 3, 2, 4, 5, 6, 7, 2, 4, 2, 1, 5, 3, 4, 9, 6, 4, 3, 4, 4, 6, 7, 6, 3, 4, 3, 6, 6, 3, 4, 4, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5]) - sample_dp_mean(samples)float32dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Mean Sample DP
Array Chunk Bytes 0.98 kiB 0.98 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type float32 numpy.ndarray - call_dosage(variants, samples)int64dask.array<chunksize=(989, 250), meta=np.ndarray>
Array Chunk Bytes 16.01 MiB 1.89 MiB Shape (8394, 250) (989, 250) Dask graph 9 chunks in 6 graph layers Data type int64 numpy.ndarray
- genotypesPandasIndex
PandasIndex(Index(['0/0', '0/1', '1/1'], dtype='object', name='genotypes'))
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
You can see that new variables have been added for variant_linreg_p_value, variant_linreg_t_value, and variant_linreg_beta.
Since sgkit doesn’t have any plotting utilities, we implement Manhattan plots and QQ plots here using Seaborn.
import seaborn as sns
%matplotlib inline
def manhattan_plot(ds):
df = ds[["variant_contig_name", "variant_contig", "variant_position", "variant_linreg_p_value"]].to_dataframe()
df["variant_linreg_log_p_value"] = -np.log10(df["variant_linreg_p_value"])
df = df.astype({"variant_position": np.int64}) # to avoid overflow in cumulative_pos
# from https://github.com/mojones/video_notebooks/blob/master/Manhattan%20plots%20in%20Python.ipynb, cell 20
running_pos = 0
cumulative_pos = []
for chrom, group_df in df.groupby("variant_contig"):
cumulative_pos.append(group_df["variant_position"] + running_pos)
running_pos += group_df["variant_position"].max()
df["cumulative_pos"] = pd.concat(cumulative_pos)
df["color group"] = df["variant_contig"].apply(lambda x : "A" if x % 2 == 0 else "B")
g = sns.relplot(
data = df,
x = "cumulative_pos",
y = "variant_linreg_log_p_value",
hue = "color group",
palette = ["blue", "orange"],
linewidth=0,
s=10,
legend=None,
aspect=3
)
g.ax.set_xlabel("Chromosome")
g.ax.set_xticks(df.groupby("variant_contig")["cumulative_pos"].median())
g.ax.set_xticklabels(df["variant_contig_name"].unique())
manhattan_plot(ds_lr)
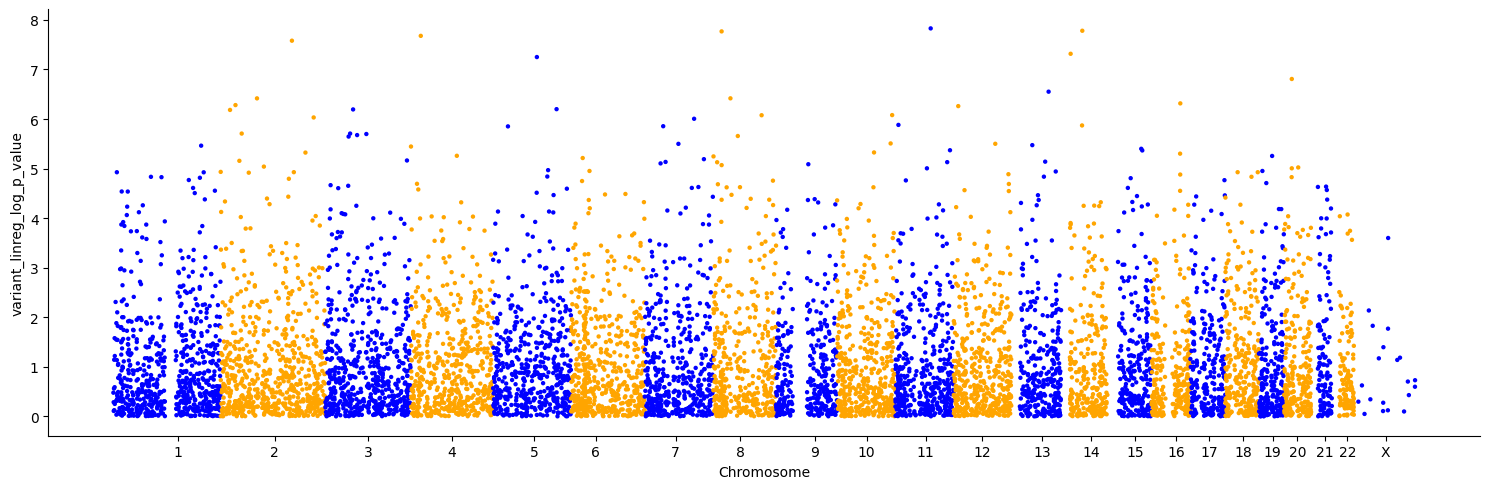
import math
import matplotlib.pyplot as plt
def qq_plot(ds):
p = ds["variant_linreg_p_value"].squeeze().values
p.sort()
n = len(p)
expected_p = -np.log10(np.arange(1, n + 1) / n)
observed_p = -np.log10(p)
max_val = math.ceil(max(np.max(expected_p), np.max(observed_p)))
df = pd.DataFrame({"Expected -log10(p)": expected_p, "Observed -log10(p)": observed_p})
fig, ax = plt.subplots(figsize=(12, 12));
g = sns.scatterplot(data=df, x="Expected -log10(p)", y="Observed -log10(p)", ax=ax, linewidth=0)
x_pred = np.linspace(0, max_val, 50)
sns.lineplot(x=x_pred, y=x_pred, ax=ax)
g.set(xlim=(0, max_val), ylim=(0, max_val))
qq_plot(ds_lr)
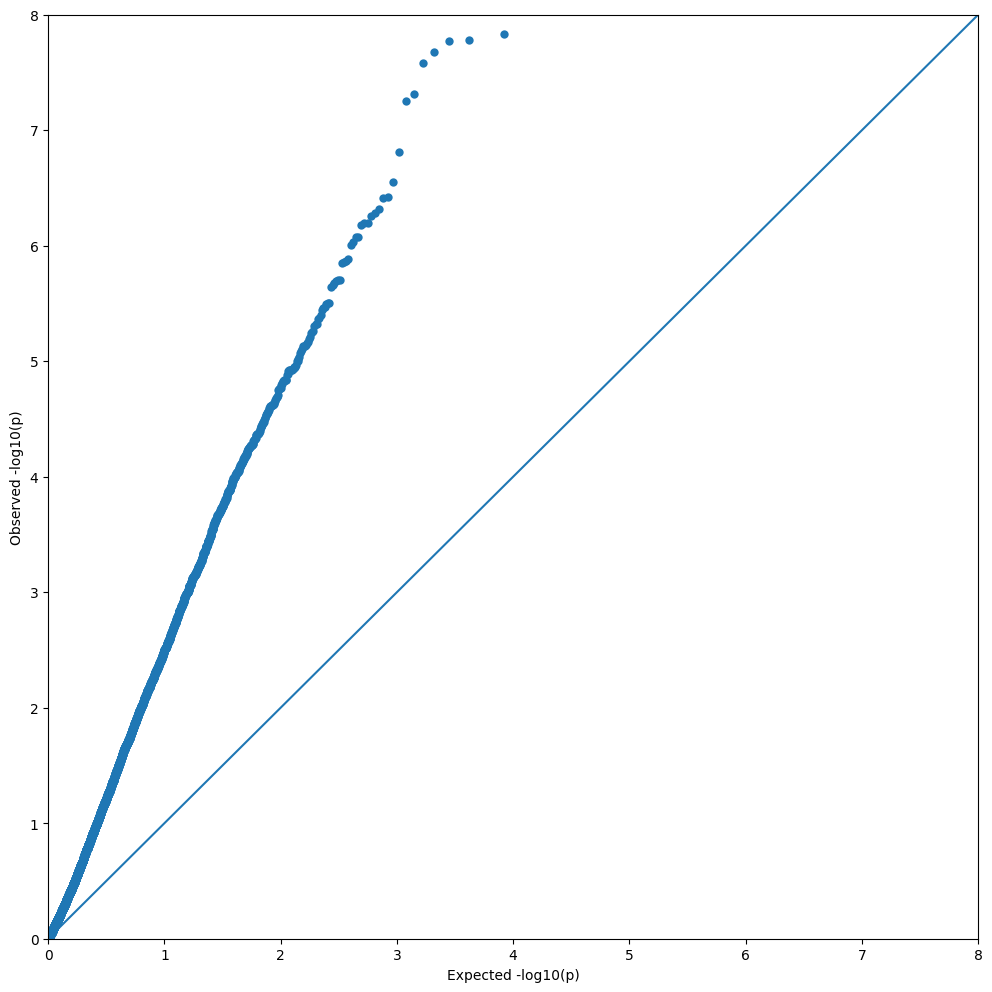
Confounded!#
As explained in the Hail tutorial, the data contains a confounder, so it is necessary to include ancestry as a covariate in the linear regression.
Rather than just use the reported ancestry, it’s better to use principal components from running a PCA on the data.
ds_pca = sg.stats.pca.count_call_alternate_alleles(ds)
# To run PCA we need to filter out variants with any missing alt allele counts
# Or where the counts are zero for all samples
variant_mask = (((ds_pca.call_alternate_allele_count < 0).any(dim="samples")) | \
(ds_pca.call_alternate_allele_count.std(dim="samples") <= 0.0)).compute()
ds_pca = ds_pca.sel(variants=~variant_mask)
ds_pca = sg.pca(ds_pca)
ds_pca.sample_pca_projection.values
array([[ -8.453593 , -26.128807 , -11.008167 , ..., -14.801194 ,
24.537308 , -1.079474 ],
[ -9.496217 , -26.319609 , -10.116498 , ..., 1.6827664 ,
7.681703 , -5.9572744 ],
[ -7.8734045 , -25.404318 , -9.85919 , ..., 4.382856 ,
-9.368463 , 6.3843317 ],
...,
[-10.974407 , -11.576627 , 20.124643 , ..., -4.4210668 ,
-0.53930056, 1.012455 ],
[-10.7544 , -11.414772 , 15.358788 , ..., 1.7951677 ,
3.4263642 , -7.985671 ],
[-13.062881 , -11.688104 , 16.351278 , ..., -7.2050548 ,
-1.7339745 , 5.17502 ]], dtype=float32)
ds_pca
<xarray.Dataset> Size: 18MB
Dimensions: (samples: 250, components: 10,
variants: 3491, genotypes: 3,
alleles: 2, ploidy: 2, contigs: 84,
filters: 1, region_index_values: 33,
region_index_fields: 6)
Coordinates:
* genotypes (genotypes) <U3 36B '0/0' '0/1' '1/1'
Dimensions without coordinates: samples, components, variants, alleles, ploidy,
contigs, filters, region_index_values,
region_index_fields
Data variables: (12/69)
sample_pca_projection (samples, components) float32 10kB dask.array<chunksize=(250, 10), meta=np.ndarray>
sample_pca_component (variants, components) float32 140kB dask.array<chunksize=(932, 10), meta=np.ndarray>
sample_pca_explained_variance (components) float32 40B dask.array<chunksize=(10,), meta=np.ndarray>
sample_pca_explained_variance_ratio (components) float32 40B dask.array<chunksize=(10,), meta=np.ndarray>
sample_pca_loading (variants, components) float32 140kB dask.array<chunksize=(932, 10), meta=np.ndarray>
call_alternate_allele_count (variants, samples) int16 2MB dask.array<chunksize=(932, 250), meta=np.ndarray>
... ...
SuperPopulation (samples) object 2kB 'EUR' ... 'SAS'
isFemale (samples) bool 250B False True ... True
PurpleHair (samples) bool 250B False ... True
CaffeineConsumption (samples) int64 2kB 4 4 4 3 ... 4 6 5 5
sample_dp_mean (samples) float32 1kB dask.array<chunksize=(250,), meta=np.ndarray>
call_dosage (variants, samples) int64 7MB dask.array<chunksize=(932, 250), meta=np.ndarray>
Attributes: (3)- samples: 250
- components: 10
- variants: 3491
- genotypes: 3
- alleles: 2
- ploidy: 2
- contigs: 84
- filters: 1
- region_index_values: 33
- region_index_fields: 6
- genotypes(genotypes)<U3'0/0' '0/1' '1/1'
array(['0/0', '0/1', '1/1'], dtype='<U3')
- sample_pca_projection(samples, components)float32dask.array<chunksize=(250, 10), meta=np.ndarray>
Array Chunk Bytes 9.77 kiB 9.77 kiB Shape (250, 10) (250, 10) Dask graph 1 chunks in 69 graph layers Data type float32 numpy.ndarray - sample_pca_component(variants, components)float32dask.array<chunksize=(932, 10), meta=np.ndarray>
Array Chunk Bytes 136.37 kiB 36.41 kiB Shape (3491, 10) (932, 10) Dask graph 4 chunks in 66 graph layers Data type float32 numpy.ndarray - sample_pca_explained_variance(components)float32dask.array<chunksize=(10,), meta=np.ndarray>
Array Chunk Bytes 40 B 40 B Shape (10,) (10,) Dask graph 1 chunks in 69 graph layers Data type float32 numpy.ndarray - sample_pca_explained_variance_ratio(components)float32dask.array<chunksize=(10,), meta=np.ndarray>
Array Chunk Bytes 40 B 40 B Shape (10,) (10,) Dask graph 1 chunks in 74 graph layers Data type float32 numpy.ndarray - sample_pca_loading(variants, components)float32dask.array<chunksize=(932, 10), meta=np.ndarray>
Array Chunk Bytes 136.37 kiB 36.41 kiB Shape (3491, 10) (932, 10) Dask graph 4 chunks in 75 graph layers Data type float32 numpy.ndarray - call_alternate_allele_count(variants, samples)int16dask.array<chunksize=(932, 250), meta=np.ndarray>
Array Chunk Bytes 1.66 MiB 455.08 kiB Shape (3491, 250) (932, 250) Dask graph 4 chunks in 18 graph layers Data type int16 numpy.ndarray - variant_hwe_p_value(variants)float64dask.array<chunksize=(932,), meta=np.ndarray>
- comment :
- P values from HWE test for each variant as float in [0, 1].
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 10 graph layers Data type float64 numpy.ndarray - variant_genotype_count(variants, genotypes)uint64dask.array<chunksize=(932, 3), meta=np.ndarray>
- comment :
- The number of observations for each possible genotype at each variant. Counts are sorted following the ordering defined in the VCF specification. - For biallelic, diploid genotypes the ordering is ``00``, ``01``, ``11`` (homozygous reference, heterozygous, homozygous alternate). - For triallelic, diploid genotypes the ordering is ``00``, ``01``, ``11``, ``02``, ``12``, ``22`` - For triallelic, triploid genotypes the ordering is ``000``, ``001``, ``011``, ``111``, ``002``, ``012``, ``112``, ``022``, ``122``, ``222``
Array Chunk Bytes 81.82 kiB 21.84 kiB Shape (3491, 3) (932, 3) Dask graph 4 chunks in 6 graph layers Data type uint64 numpy.ndarray - genotype_id(genotypes)<U3dask.array<chunksize=(3,), meta=np.ndarray>
- comment :
- VCF style genotype strings for all possible genotypes given the size of the ploidy and alleles dimensions. The ordering of genotype strings follows the ordering outlined in the VCF specification for arrays of size "G".
Array Chunk Bytes 36 B 36 B Shape (3,) (3,) Dask graph 1 chunks in 4 graph layers Data type - variant_n_called(variants)int64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 10 graph layers Data type int64 numpy.ndarray - variant_call_rate(variants)float64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 12 graph layers Data type float64 numpy.ndarray - variant_n_het(variants)int64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 9 graph layers Data type int64 numpy.ndarray - variant_n_hom_ref(variants)int64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 9 graph layers Data type int64 numpy.ndarray - variant_n_hom_alt(variants)int64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 9 graph layers Data type int64 numpy.ndarray - variant_n_non_ref(variants)int64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 12 graph layers Data type int64 numpy.ndarray - variant_allele_count(variants, alleles)uint64dask.array<chunksize=(932, 2), meta=np.ndarray>
- comment :
- Variant allele counts. With shape (variants, alleles) and values corresponding to the number of non-missing occurrences of each allele.
Array Chunk Bytes 54.55 kiB 14.56 kiB Shape (3491, 2) (932, 2) Dask graph 4 chunks in 7 graph layers Data type uint64 numpy.ndarray - variant_allele_total(variants)int64dask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 10 graph layers Data type int64 numpy.ndarray - variant_allele_frequency(variants, alleles)float64dask.array<chunksize=(932, 2), meta=np.ndarray>
Array Chunk Bytes 54.55 kiB 14.56 kiB Shape (3491, 2) (932, 2) Dask graph 4 chunks in 15 graph layers Data type float64 numpy.ndarray - sample_n_called(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 10 graph layers Data type int64 numpy.ndarray - sample_call_rate(samples)float64dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Sample call rate
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type float64 numpy.ndarray - sample_n_het(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_hom_alt(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type int64 numpy.ndarray - sample_n_non_ref(samples)int64dask.array<chunksize=(250,), meta=np.ndarray>
Array Chunk Bytes 1.95 kiB 1.95 kiB Shape (250,) (250,) Dask graph 1 chunks in 12 graph layers Data type int64 numpy.ndarray - call_AD(variants, samples, alleles)int8dask.array<chunksize=(932, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 1.66 MiB 455.08 kiB Shape (3491, 250, 2) (932, 250, 2) Dask graph 4 chunks in 5 graph layers Data type int8 numpy.ndarray - call_DP(variants, samples)int8dask.array<chunksize=(932, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 852.29 kiB 227.54 kiB Shape (3491, 250) (932, 250) Dask graph 4 chunks in 5 graph layers Data type int8 numpy.ndarray - call_GQ(variants, samples)int8dask.array<chunksize=(932, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 852.29 kiB 227.54 kiB Shape (3491, 250) (932, 250) Dask graph 4 chunks in 5 graph layers Data type int8 numpy.ndarray - call_genotype(variants, samples, ploidy)int8dask.array<chunksize=(932, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 1.66 MiB 455.08 kiB Shape (3491, 250, 2) (932, 250, 2) Dask graph 4 chunks in 5 graph layers Data type int8 numpy.ndarray - call_genotype_mask(variants, samples, ploidy)booldask.array<chunksize=(932, 250, 2), meta=np.ndarray>
- description :
Array Chunk Bytes 1.66 MiB 455.08 kiB Shape (3491, 250, 2) (932, 250, 2) Dask graph 4 chunks in 5 graph layers Data type bool numpy.ndarray - call_genotype_phased(variants, samples)booldask.array<chunksize=(932, 250), meta=np.ndarray>
- description :
Array Chunk Bytes 852.29 kiB 227.54 kiB Shape (3491, 250) (932, 250) Dask graph 4 chunks in 5 graph layers Data type bool numpy.ndarray - contig_id(contigs)objectdask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - contig_length(contigs)float64dask.array<chunksize=(84,), meta=np.ndarray>
Array Chunk Bytes 672 B 672 B Shape (84,) (84,) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - filter_description(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - filter_id(filters)objectdask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 8 B 8 B Shape (1,) (1,) Dask graph 1 chunks in 2 graph layers Data type object numpy.ndarray - region_index(region_index_values, region_index_fields)int32dask.array<chunksize=(33, 6), meta=np.ndarray>
Array Chunk Bytes 792 B 792 B Shape (33, 6) (33, 6) Dask graph 1 chunks in 2 graph layers Data type int32 numpy.ndarray - variant_AC(variants)int16dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 6.82 kiB 1.82 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int16 numpy.ndarray - variant_AF(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_AN(variants)int16dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 6.82 kiB 1.82 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int16 numpy.ndarray - variant_BaseQRankSum(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_ClippingRankSum(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_DP(variants)int32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int32 numpy.ndarray - variant_DS(variants)booldask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 3.41 kiB 0.91 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type bool numpy.ndarray - variant_FS(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_HaplotypeScore(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_InbreedingCoeff(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_MLEAC(variants)int16dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 6.82 kiB 1.82 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int16 numpy.ndarray - variant_MLEAF(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_MQ(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_MQ0(variants)int8dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 3.41 kiB 0.91 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int8 numpy.ndarray - variant_MQRankSum(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_QD(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_ReadPosRankSum(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_allele(variants, alleles)objectdask.array<chunksize=(932, 2), meta=np.ndarray>
- description :
- List of the reference and alternate alleles
Array Chunk Bytes 54.55 kiB 14.56 kiB Shape (3491, 2) (932, 2) Dask graph 4 chunks in 4 graph layers Data type object numpy.ndarray - variant_contig(variants)int8dask.array<chunksize=(932,), meta=np.ndarray>
- description :
- An identifier from the reference genome or an angle-bracketed ID string pointing to a contig in the assembly file
Array Chunk Bytes 3.41 kiB 0.91 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int8 numpy.ndarray - variant_filter(variants, filters)booldask.array<chunksize=(932, 1), meta=np.ndarray>
- description :
- Filter status of the variant
Array Chunk Bytes 3.41 kiB 0.91 kiB Shape (3491, 1) (932, 1) Dask graph 4 chunks in 4 graph layers Data type bool numpy.ndarray - variant_id(variants)objectdask.array<chunksize=(932,), meta=np.ndarray>
- description :
- List of unique identifiers where applicable
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type object numpy.ndarray - variant_id_mask(variants)booldask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 3.41 kiB 0.91 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type bool numpy.ndarray - variant_length(variants)int8dask.array<chunksize=(932,), meta=np.ndarray>
- description :
- The length of the variant measured in bases
Array Chunk Bytes 3.41 kiB 0.91 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int8 numpy.ndarray - variant_position(variants)int32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
- The reference position
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type int32 numpy.ndarray - variant_quality(variants)float32dask.array<chunksize=(932,), meta=np.ndarray>
- description :
- Phred-scaled quality score
Array Chunk Bytes 13.64 kiB 3.64 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type float32 numpy.ndarray - variant_set(variants)objectdask.array<chunksize=(932,), meta=np.ndarray>
- description :
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 4 graph layers Data type object numpy.ndarray - variant_contig_name(variants)objectdask.array<chunksize=(932,), meta=np.ndarray>
Array Chunk Bytes 27.27 kiB 7.28 kiB Shape (3491,) (932,) Dask graph 4 chunks in 10 graph layers Data type object numpy.ndarray - Population(samples)object'GBR' 'GBR' 'GBR' ... 'GIH' 'GIH'
array(['GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'GBR', 'FIN', 'FIN', 'FIN', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'CHS', 'PUR', 'CDX', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'PUR', 'CLM', 'CLM', 'CLM', 'GBR', 'CLM', 'PUR', 'CLM', 'CLM', 'CLM', 'IBS', 'PEL', 'IBS', 'IBS', 'IBS', 'IBS', 'IBS', 'GBR', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'ACB', 'PEL', 'PEL', 'PEL', 'PEL', 'ACB', 'KHV', 'ACB', 'KHV', 'KHV', 'KHV', 'KHV', 'KHV', 'CDX', 'CDX', 'CDX', 'IBS', 'IBS', 'CDX', 'PEL', 'PEL', 'PEL', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'CDX', 'ACB', 'GWD', 'GWD', 'ACB', 'KHV', 'GWD', 'GWD', 'ACB', 'GWD', 'PJL', 'GWD', 'PJL', 'PJL', 'PJL', 'PJL', 'PJL', 'GWD', 'GWD', 'GWD', 'PJL', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'BEB', 'GWD', 'MSL', 'MSL', 'ESN', 'ESN', 'ESN', 'MSL', 'PJL', 'GWD', 'GWD', 'GWD', 'ESN', 'ESN', 'ESN', 'ESN', 'MSL', 'MSL', 'MSL', 'MSL', 'PJL', 'PJL', 'ESN', 'MSL', 'MSL', 'BEB', 'BEB', 'BEB', 'PJL', 'STU', 'STU', 'STU', 'ITU', 'STU', 'STU', 'BEB', 'BEB', 'BEB', 'STU', 'ITU', 'STU', 'BEB', 'BEB', 'STU', 'ITU', 'ITU', 'ITU', 'ITU', 'STU', 'BEB', 'BEB', 'ITU', 'STU', 'STU', 'ITU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'CEU', 'YRI', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'CHB', 'YRI', 'YRI', 'YRI', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'JPT', 'YRI', 'YRI', 'YRI', 'YRI', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'LWK', 'MXL', 'MXL', 'MXL', 'MXL', 'MXL', 'ASW', 'MXL', 'MXL', 'MXL', 'ASW', 'ASW', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'TSI', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH', 'GIH'], dtype=object) - SuperPopulation(samples)object'EUR' 'EUR' 'EUR' ... 'SAS' 'SAS'
array(['EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AMR', 'EAS', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'EUR', 'AMR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'EAS', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EUR', 'EUR', 'EAS', 'AMR', 'AMR', 'AMR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'AFR', 'AFR', 'AFR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'EAS', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AFR', 'AMR', 'AMR', 'AMR', 'AMR', 'AMR', 'AFR', 'AMR', 'AMR', 'AMR', 'AFR', 'AFR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'EUR', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS', 'SAS'], dtype=object) - isFemale(samples)boolFalse True False ... False True
array([False, True, False, True, False, False, True, False, True, False, False, True, False, True, False, False, False, True, True, True, False, True, True, False, True, True, False, False, True, False, True, False, True, True, False, True, True, False, False, True, True, True, False, True, False, False, True, True, True, True, True, True, True, True, True, False, True, True, True, True, True, True, False, False, True, False, True, True, True, False, False, True, True, True, True, True, False, False, False, True, True, False, False, False, False, False, False, False, False, False, True, True, False, True, False, False, True, True, True, False, True, False, True, True, False, False, True, False, False, False, False, True, True, True, True, False, True, False, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, True, True, False, True, False, True, True, False, True, True, False, True, False, True, False, True, True, False, False, False, False, True, False, True, True, False, True, True, True, True, True, False, True, False, True, True, False, False, False, False, True, False, False, True, False, True, False, False, True, False, True, False, True, True, False, True, False, False, True, True, False, False, True, True, True, False, False, False, False, True, False, False, True, True, True, False, True, False, True, False, True, True, True, False, True, False, True, False, True, False, False, True, False, False, True, False, True, True, True, False, True, False, True, False, False, True, True, True, True, True, False, False, False, False, False, True]) - PurpleHair(samples)boolFalse False False ... True True
array([False, False, False, False, False, True, False, False, True, True, False, False, True, True, True, True, True, True, True, False, True, True, False, False, False, False, False, True, False, True, True, True, False, True, False, True, False, False, False, False, True, False, False, True, False, False, True, True, False, True, False, False, False, False, False, True, True, False, False, True, True, True, False, False, True, False, True, True, True, True, True, True, True, True, False, True, True, True, True, True, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, True, True, True, True, True, False, True, False, False, False, True, True, False, False, True, True, True, True, True, False, False, True, True, False, False, False, False, False, True, False, False, False, False, True, False, False, False, False, False, False, False, False, True, False, False, True, True, True, False, True, False, True, True, True, True, True, True, False, False, False, False, False, False, True, True, True, True, False, False, True, False, True, True, False, False, True, False, False, True, True, True, True, True, True, False, True, True, False, False, True, True, True, False, True, False, False, False, True, True, True, False, False, False, False, False, True, False, False, False, True, False, False, True, False, False, False, False, False, True, False, True, True, False, False, False, False, True, False, True, False, True, True, False, False, False, False, True, True, True, True, True, True, False, False, True, False, False, True, True, True, False, True, True]) - CaffeineConsumption(samples)int644 4 4 3 6 2 2 5 ... 5 6 4 6 4 6 5 5
array([4, 4, 4, 3, 6, 2, 2, 5, 5, 4, 6, 5, 7, 5, 7, 1, 5, 5, 4, 4, 5, 5, 5, 6, 4, 4, 6, 3, 3, 5, 4, 4, 5, 5, 4, 6, 5, 4, 4, 5, 6, 3, 7, 5, 5, 6, 3, 2, 5, 5, 4, 6, 5, 6, 4, 6, 7, 6, 7, 3, 5, 6, 5, 6, 4, 5, 4, 4, 5, 8, 3, 4, 4, 7, 5, 2, 6, 7, 6, 5, 3, 4, 5, 5, 5, 5, 6, 4, 5, 7, 2, 3, 3, 2, 6, 4, 2, 6, 5, 3, 4, 7, 6, 7, 6, 3, 4, 2, 2, 5, 6, 7, 8, 6, 2, 3, 2, 0, 5, 7, 5, 1, 4, 3, 2, 4, 6, 5, 4, 4, 1, 5, 5, 3, 1, 1, 3, 2, 4, 2, 1, 3, 3, 4, 4, 5, 6, 5, 4, 5, 0, 4, 5, 4, 3, 3, 4, 4, 3, 5, 6, 5, 3, 4, 4, 6, 3, 5, 5, 4, 5, 3, 5, 4, 6, 5, 7, 5, 6, 6, 4, 4, 5, 3, 5, 4, 3, 8, 4, 6, 8, 3, 4, 2, 5, 6, 6, 4, 3, 5, 7, 4, 5, 5, 6, 3, 2, 4, 5, 6, 7, 2, 4, 2, 1, 5, 3, 4, 9, 6, 4, 3, 4, 4, 6, 7, 6, 3, 4, 3, 6, 6, 3, 4, 4, 4, 6, 7, 4, 5, 4, 5, 5, 6, 4, 6, 4, 6, 5, 5]) - sample_dp_mean(samples)float32dask.array<chunksize=(250,), meta=np.ndarray>
- long_name :
- Mean Sample DP
Array Chunk Bytes 0.98 kiB 0.98 kiB Shape (250,) (250,) Dask graph 1 chunks in 9 graph layers Data type float32 numpy.ndarray - call_dosage(variants, samples)int64dask.array<chunksize=(932, 250), meta=np.ndarray>
Array Chunk Bytes 6.66 MiB 1.78 MiB Shape (3491, 250) (932, 250) Dask graph 4 chunks in 7 graph layers Data type int64 numpy.ndarray
- genotypesPandasIndex
PandasIndex(Index(['0/0', '0/1', '1/1'], dtype='object', name='genotypes'))
- source :
- bio2zarr-0.1.6
- vcf_meta_information :
- [['fileformat', 'VCFv4.2'], ['hailversion', '0.2-29fbaeaf265e']]
- vcf_zarr_version :
- 0.4
Let’s plot the first two components. Notice how they cluster by ancestry.
ds_pca["sample_pca_projection_0"] = ds_pca.sample_pca_projection[:,0]
ds_pca["sample_pca_projection_1"] = ds_pca.sample_pca_projection[:,1]
ds_pca["sample_pca_projection_2"] = ds_pca.sample_pca_projection[:,2]
# Following does not work with recent versions of xarray, see https://github.com/sgkit-dev/sgkit/issues/934
#ds_pca.plot.scatter(x="sample_pca_projection_0", y="sample_pca_projection_1", hue="SuperPopulation", size=8, s=10);
Now we can rerun our linear regression, controlling for sample sex and the first few principal components.
# copy pca components back to dataset with full set of variants to run linear regression again
ds["sample_pca_projection_0"] = ds_pca.sample_pca_projection[:,0]
ds["sample_pca_projection_1"] = ds_pca.sample_pca_projection[:,1]
ds["sample_pca_projection_2"] = ds_pca.sample_pca_projection[:,2]
ds_lr = sg.gwas_linear_regression(ds, dosage="call_dosage",
add_intercept=True,
covariates=["isFemale", "sample_pca_projection_0", "sample_pca_projection_1", "sample_pca_projection_2"],
traits=["CaffeineConsumption"])
qq_plot(ds_lr)
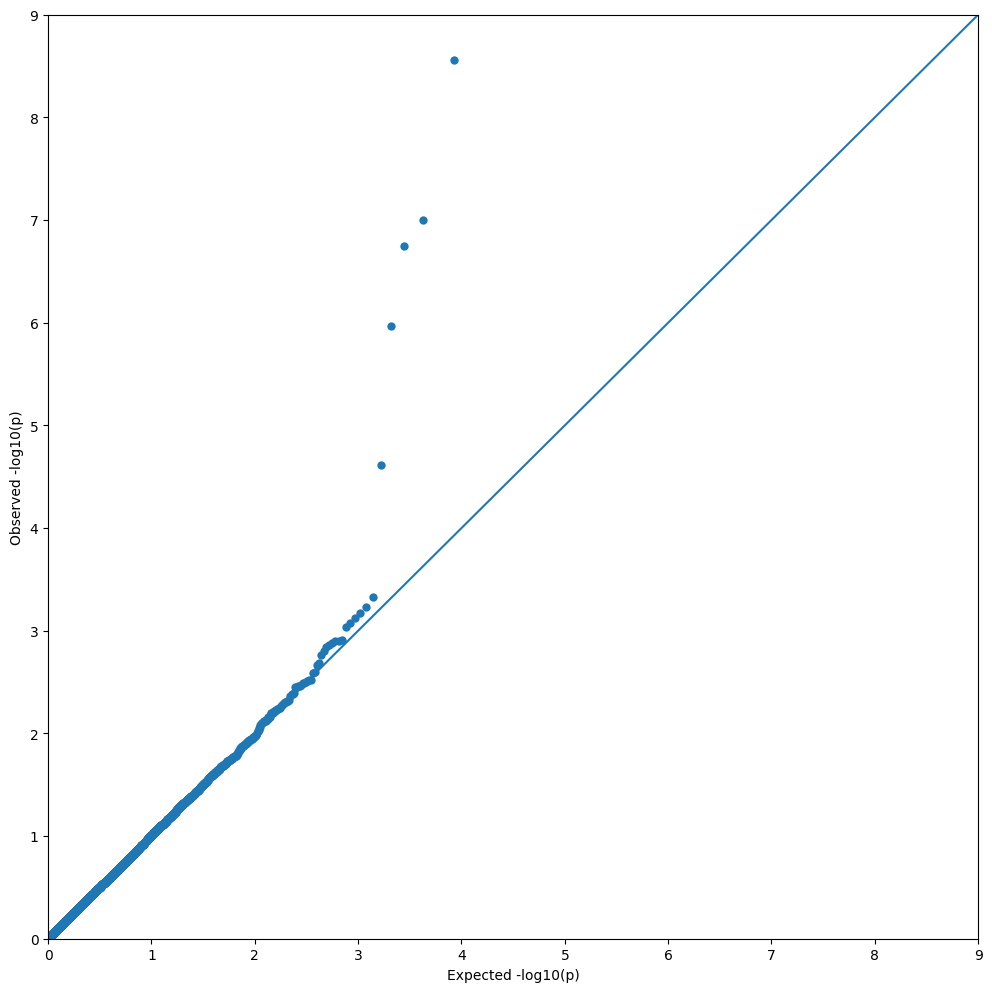
manhattan_plot(ds_lr)
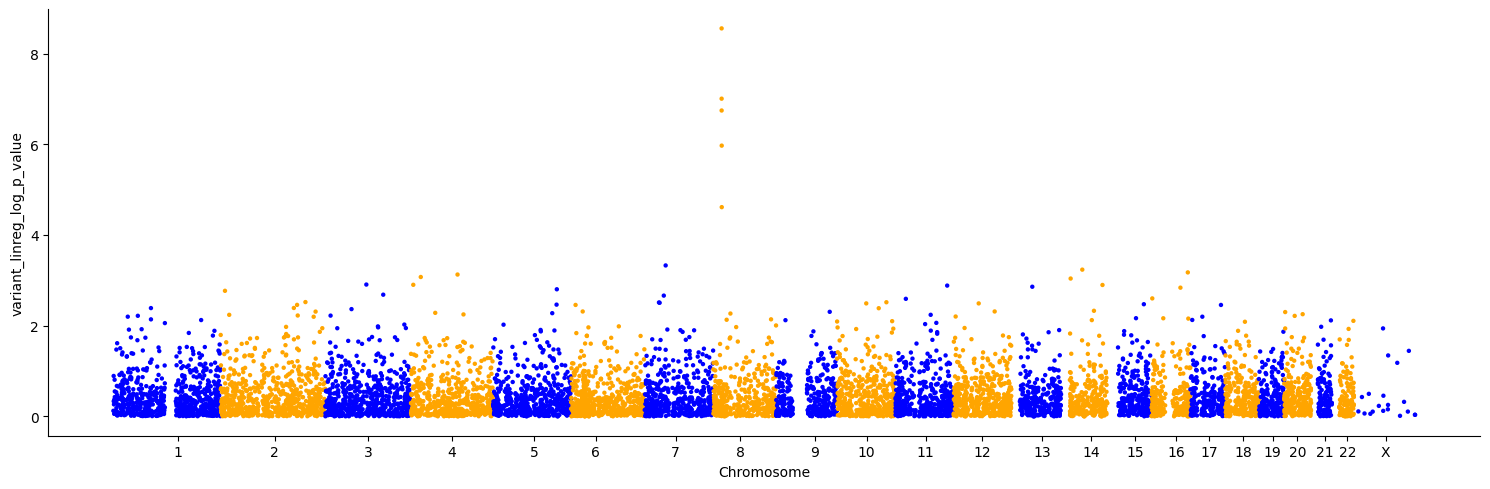
The “caffeine consumption” locus in chromosome 8 is clearly apparent, just like in the Hail tutorial.

